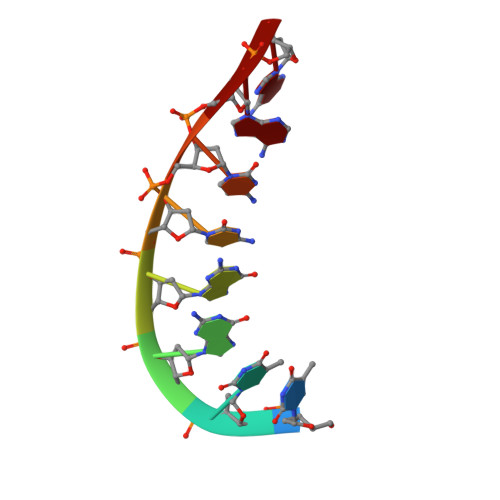
A high-resolution structure of a DNA-chromomycin-Co(II) complex determined from pseudocontact shifts in nuclear magnetic resonance.
Gochin, M.(2000) Structure 8: 441-452
- PubMed: 10801486
- DOI: https://doi.org/10.1016/s0969-2126(00)00124-6
- Primary Citation of Related Structures:
1EKH, 1EKI - PubMed Abstract:
The drug chromomycin-A(3) binds to the minor groove of DNA and requires a divalent metal ion for complex formation. (1)H, (31)P and (13)C pseudocontact shifts occurring in the presence of a tightly bound divalent cobalt ion in the complex between d(TTGGCCAA)(2) and chromomycin-A(3) have been used to determine the structure of the complex. The accuracy of the structure was verified by validation with nuclear Overhauser enhancements (NOEs) and J-coupling constants not used in the structure calculation.
Organizational Affiliation:
Department of Microbiology, University of the Pacific School of Dentistry, San Francisco, CA 94115, USA. miriam@picasso. ucsf.edu