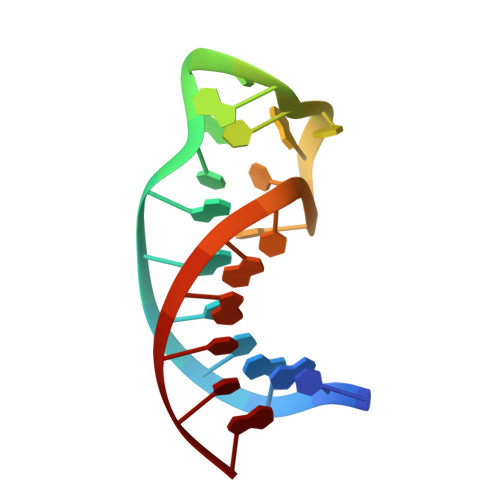
Structure determination and binding kinetics of a DNA aptamer-argininamide complex.
Robertson, S.A., Harada, K., Frankel, A.D., Wemmer, D.E.(2000) Biochemistry 39: 946-954
- PubMed: 10653638
- DOI: https://doi.org/10.1021/bi9915061
- Primary Citation of Related Structures:
1DB6 - PubMed Abstract:
The structure of a DNA aptamer, which was selected for specific binding to arginine, was determined using NMR spectroscopy. The sequence forms a hairpin loop, with residues important for binding occurring in the loop region. Binding of argininamide induces formation of one Watson-Crick and two non-Watson-Crick base pairs, which facilitate generation of a binding pocket. The specificity for arginine seems to arise from contacts between the guanidino end of the arginine and phosphates, with atoms positioned by the shape of the pocket. Complex binding kinetics are observed suggesting that there is a slow interconversion of two forms of the DNA, which have different binding affinities. These data provide information on the process of adaptive recognition of a ligand by an aptamer.
Organizational Affiliation:
Department of Chemistry, University of California, Berkeley, California 94720-1460, USA.