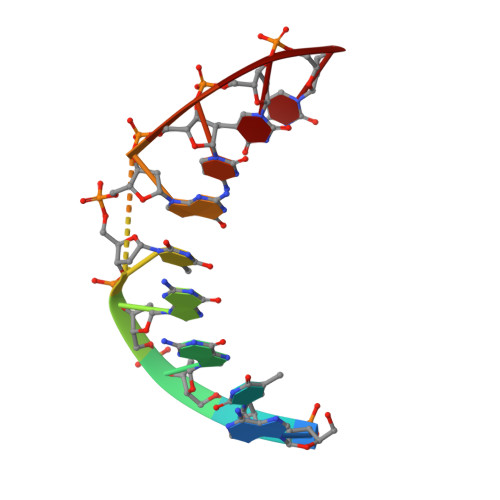
Solution structure of a DNA.RNA hybrid containing an alpha-anomeric thymidine and polarity reversals: d(ATGG-3'-3'-alphaT-5'-5'-GCTC). r(gagcaccau).
Aramini, J.M., Germann, M.W.(1999) Biochemistry 38: 15448-15458
- PubMed: 10569927
- DOI: https://doi.org/10.1021/bi9915418
- Primary Citation of Related Structures:
1C2Q - PubMed Abstract:
We report the thermodynamic and structural properties of an alpha-containing DNA.RNA nonamer hybrid duplex, d(ATGG-3'-3'-alphaT-5'-5'-GCTC).r(gagcaccau). The RNA strand corresponds to the core of the initiation sequence for the transcript of the erbB-2 oncogene. The tandem anomeric and polarity changes in the DNA strand result in a slight decrease in thermostability (DeltaT(m) = -2.8 degrees C) compared to the unmodified control hybrid. The three-dimensional solution structure determination of the alpha-containing DNA.RNA hybrid, conducted via restrained molecular dynamics using interproton distance (nuclear Overhauser enhancement) and furanose ring torsion angle (J-based) restraints, converged to a final ensemble of structures from unique starting models. In agreement with hyperchromicity and circular dichroism data, the final average structure derived from this ensemble is consistent with an overall A-like motif featuring Watson-Crick base pairing and base stacking across the entire sequence, albeit with localized B-like traits within the DNA strand. Comparative pseudorotation analyses of the J-coupling data for this hybrid and its unmodified control reveal a surprising increase in S-puckering for two nucleotides immediately upstream of the 3'-3' linkage, and the associated narrowing of the minor groove in this portion of the hybrid. Other structural perturbations are localized to and diagnostic of the central alpha-nucleotide and juxtaposed polarity reversals. The structural information presented here has direct relevance to the design of future antisense oligonucleotides composed of these modifications.
Organizational Affiliation:
Department of Microbiology and Immunology, Kimmel Cancer Institute, Thomas Jefferson University, Philadelphia, Pennsylvania 19107, USA.