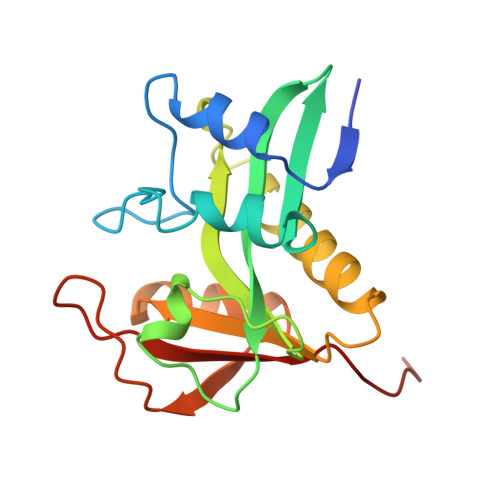
Melatonin biosynthesis: the structure of serotonin N-acetyltransferase at 2.5 A resolution suggests a catalytic mechanism.
Hickman, A.B., Klein, D.C., Dyda, F.(1999) Mol Cell 3: 23-32
- PubMed: 10024876
- DOI: https://doi.org/10.1016/s1097-2765(00)80171-9
- Primary Citation of Related Structures:
1B6B - PubMed Abstract:
Conversion of serotonin to N-acetylserotonin, the precursor of the circadian neurohormone melatonin, is catalyzed by serotonin N-acetyltransferase (AANAT) in a reaction requiring acetyl coenzyme A (AcCoA). AANAT is a globular protein consisting of an eight-stranded beta sheet flanked by five alpha helices; a conserved motif in the center of the beta sheet forms the cofactor binding site. Three polypeptide loops converge above the AcCoA binding site, creating a hydrophobic funnel leading toward the cofactor and serotonin binding sites in the protein interior. Two conserved histidines not found in other NATs are located at the bottom of the funnel in the active site, suggesting a catalytic mechanism for acetylation involving imidazole groups acting as general acid/base catalysts.
Organizational Affiliation:
Laboratory of Developmental Neurobiology, National Institute of Child Health and Human Development, Bethesda, Maryland 20892, USA.