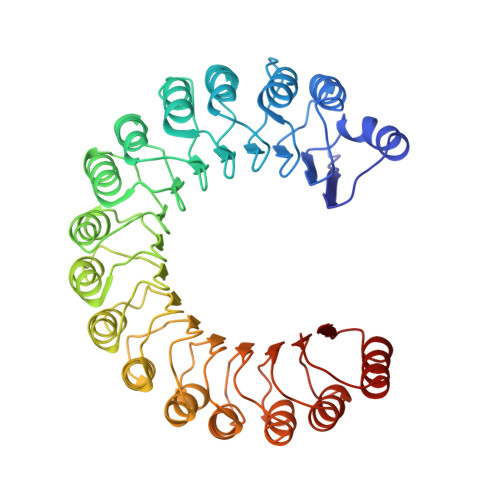
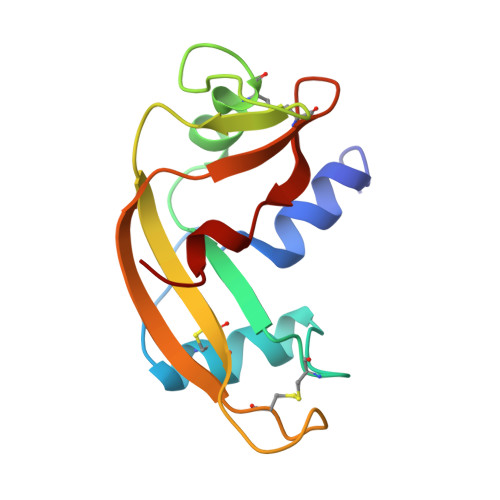
Molecular recognition of human angiogenin by placental ribonuclease inhibitor--an X-ray crystallographic study at 2.0 A resolution.
Papageorgiou, A.C., Shapiro, R., Acharya, K.R.(1997) EMBO J 16: 5162-5177
- PubMed: 9311977
- DOI: https://doi.org/10.1093/emboj/16.17.5162
- Primary Citation of Related Structures:
1A4Y - PubMed Abstract:
Human placental RNase inhibitor (hRI), a leucine-rich repeat protein, binds the blood vessel-inducing protein human angiogenin (Ang) with extraordinary affinity (Ki <1 fM). Here we report a 2.0 A resolution crystal structure for the hRI-Ang complex that, together with extensive mutagenesis data from earlier studies, reveals the molecular features of this tight interaction. The hRI-Ang binding interface is large and encompasses 26 residues from hRI and 24 from Ang, recruited from multiple domains of both proteins. However, a substantial fraction of the energetically important contacts involve only a single region of each: the C-terminal segment 434-460 of hRI and the ribonucleolytic active centre of Ang, most notably the catalytic residue Lys40. Although the overall docking of Ang resembles that observed for RNase A in the crystal structure of its complex with the porcine RNase inhibitor, the vast majority of the interactions in the two complexes are distinctive, indicating that the broad specificity of the inhibitor for pancreatic RNase superfamily proteins is based largely on its capacity to recognize features unique to each of them. The implications of these findings for the development of small, hRI-based inhibitors of Ang for therapeutic use are discussed.
Organizational Affiliation:
Department of Biology and Biochemistry, University of Bath, Claverton Down, Bath BA2 7AY, UK.