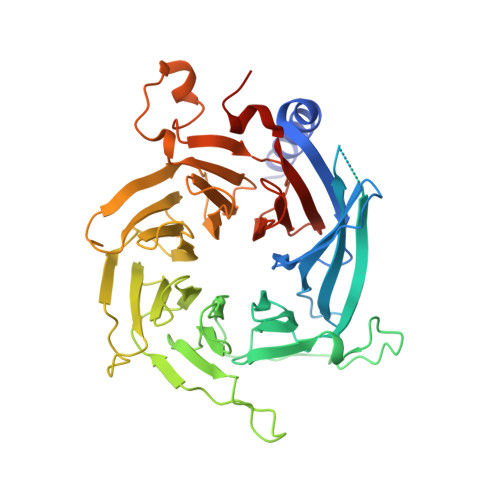
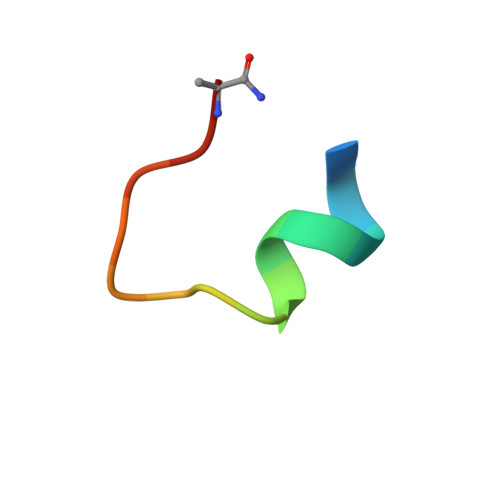
Structure Based Design of Bicyclic Peptide Inhibitors of RbAp48.
Hart, P.'., Hommen, P., Noisier, A., Krzyzanowski, A., Schuler, D., Porfetye, A.T., Akbarzadeh, M., Vetter, I.R., Adihou, H., Waldmann, H.(2021) Angew Chem Int Ed Engl 60: 1813-1820
- PubMed: 33022847
- DOI: https://doi.org/10.1002/anie.202009749
- Primary Citation of Related Structures:
6ZRC, 6ZRD - PubMed Abstract:
The scaffolding protein RbAp48 is part of several epigenetic regulation complexes and is overexpressed in a variety of cancers. In order to develop tool compounds for the study of RbAp48 function, we have developed peptide inhibitors targeting the protein-protein interaction interface between RbAp48 and the scaffold protein MTA1. Based on a MTA1-derived linear peptide with low micromolar affinity and informed by crystallographic analysis, a bicyclic peptide was developed that inhibits the RbAp48/MTA1 interaction with a very low nanomolar K D value of 8.56 nM, and which showed appreciable stability against cellular proteases. Design included exchange of a polar amide cyclization strategy to hydrophobic aromatic linkers enabling mono- and bicyclization by means of cysteine alkylation, which improved affinity by direct interaction of the linkers with a hydrophobic residue on RbAp48. Our results demonstrate that stepwise evolution of a structure-based design is a suitable strategy for inhibitor development targeting PPIs.
Organizational Affiliation:
Department of Chemical Biology, Max Planck Institute of Molecular Physiology, Otto-Hahn-Strasse 11, 44227, Dortmund, Germany.