Structural insights into the Ca 2+ -dependent gating of the human mitochondrial calcium uniporter.
Wang, Y., Han, Y., She, J., Nguyen, N.X., Mootha, V.K., Bai, X.C., Jiang, Y.(2020) Elife 9
- PubMed: 32762847
- DOI: https://doi.org/10.7554/eLife.60513
- Primary Citation of Related Structures:
6XJV, 6XJX - PubMed Abstract:
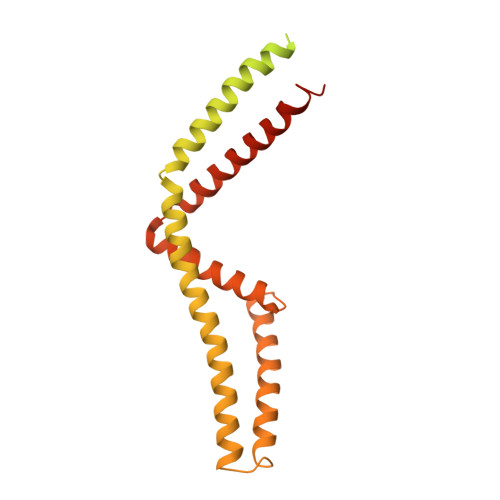
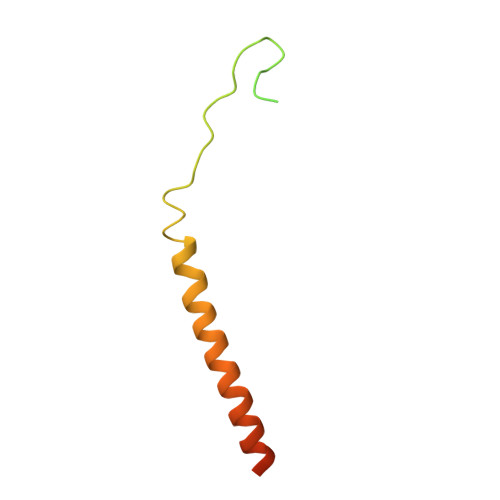
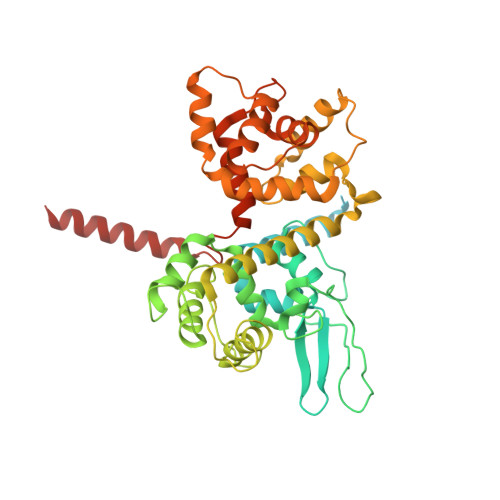
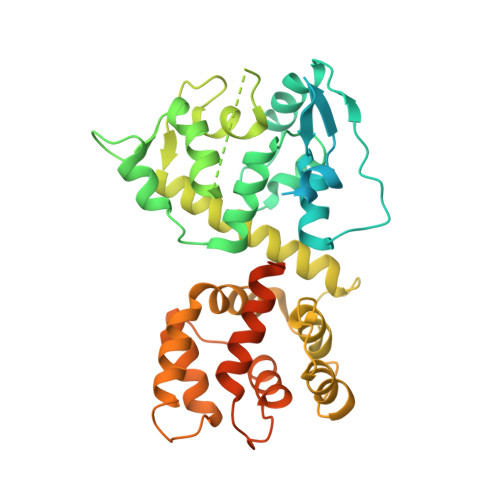
Mitochondrial Ca 2+ uptake is mediated by an inner mitochondrial membrane protein called the mitochondrial calcium uniporter. In humans, the uniporter functions as a holocomplex consisting of MCU, EMRE, MICU1 and MICU2, among which MCU and EMRE form a subcomplex and function as the conductive channel while MICU1 and MICU2 are EF-hand proteins that regulate the channel activity in a Ca 2+ -dependent manner. Here, we present the EM structures of the human mitochondrial calcium uniporter holocomplex (uniplex) in the presence and absence of Ca 2+ , revealing distinct Ca 2+ dependent assembly of the uniplex. Our structural observations suggest that Ca 2+ changes the dimerization interaction between MICU1 and MICU2, which in turn determines how the MICU1-MICU2 subcomplex interacts with the MCU-EMRE channel and, consequently, changes the distribution of the uniplex assemblies between the blocked and unblocked states.
Organizational Affiliation:
Department of Physiology, University of Texas Southwestern Medical Center, Dallas, United States.