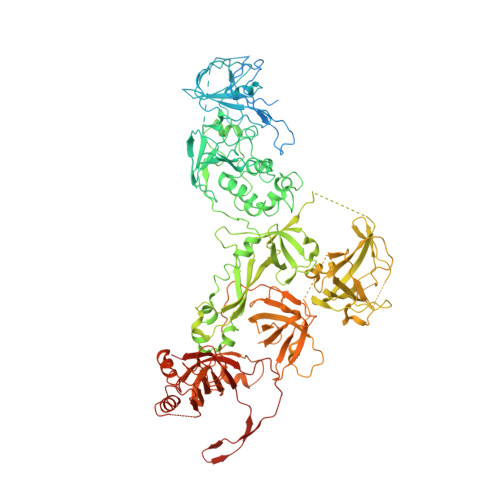
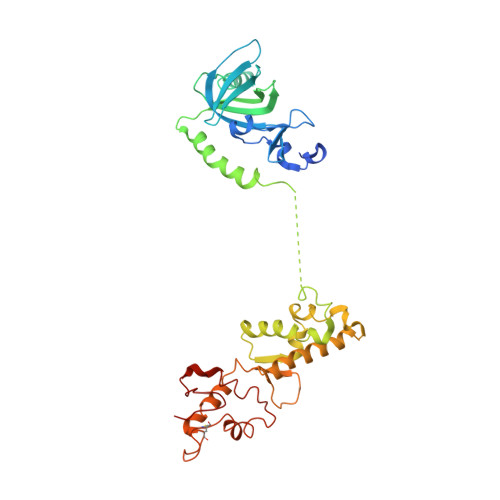
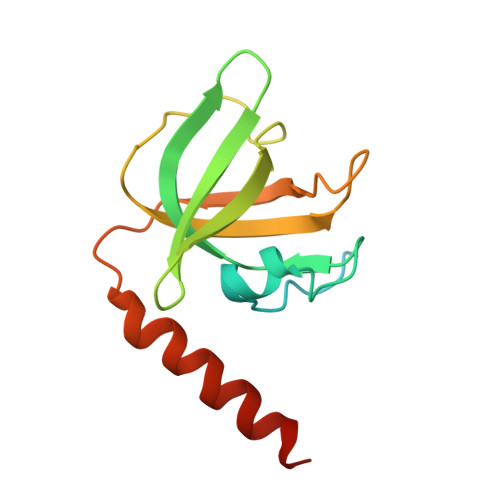
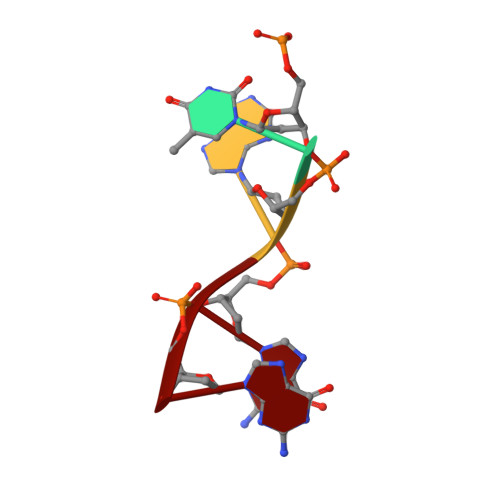
The structure of human CST reveals a decameric assembly bound to telomeric DNA.
Lim, C.J., Barbour, A.T., Zaug, A.J., Goodrich, K.J., McKay, A.E., Wuttke, D.S., Cech, T.R.(2020) Science 368: 1081-1085
- PubMed: 32499435
- DOI: https://doi.org/10.1126/science.aaz9649
- Primary Citation of Related Structures:
6W6W - PubMed Abstract:
The CTC1-STN1-TEN1 (CST) complex is essential for telomere maintenance and resolution of stalled replication forks genome-wide. Here, we report the 3.0-angstrom cryo-electron microscopy structure of human CST bound to telomeric single-stranded DNA (ssDNA), which assembles as a decameric supercomplex. The atomic model of the 134-kilodalton CTC1 subunit, built almost entirely de novo, reveals the overall architecture of CST and the DNA-binding anchor site. The carboxyl-terminal domain of STN1 interacts with CTC1 at two separate docking sites, allowing allosteric mediation of CST decamer assembly. Furthermore, ssDNA appears to staple two monomers to nucleate decamer assembly. CTC1 has stronger structural similarity to Replication Protein A than the expected similarity to yeast Cdc13. The decameric structure suggests that CST can organize ssDNA analogously to the nucleosome's organization of double-stranded DNA.
Organizational Affiliation:
Department of Biochemistry, University of Colorado Boulder, Boulder, CO 80303, USA.