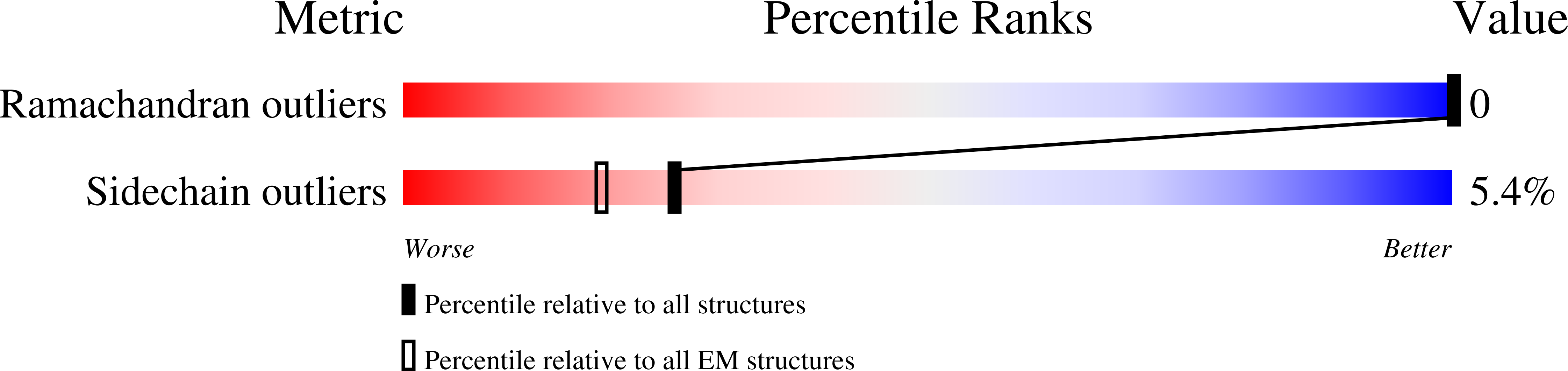Amphipathic Motifs Regulate N-BAR Protein Endophilin B1 Auto-inhibition and Drive Membrane Remodeling.
Bhatt, V.S., Ashley, R., Sundborger-Lunna, A.(2021) Structure 29: 61
- PubMed: 33086035
- DOI: https://doi.org/10.1016/j.str.2020.09.012
- Primary Citation of Related Structures:
6UP6, 6UPN - PubMed Abstract:
Membrane remodeling is a common theme in a variety of cellular processes. Here, we investigated membrane remodeling N-BAR protein endophilin B1, a critical player in diverse intracellular trafficking events, including mitochondrial and Golgi fission, and apoptosis. We find that endophilin B1 assembles into helical scaffolds on membranes, and that both membrane binding and assembly are driven by interactions between N-terminal helix H0 and the lipid bilayer. Furthermore, we find that endophilin B1 membrane remodeling is auto-inhibited and identify direct SH3 domain-H0 interactions as the underlying mechanism. Our results indicate that lipid composition plays a role in dictating endophilin B1 activity. Taken together, this study provides insight into a poorly understood N-BAR protein family member and highlights molecular mechanisms that may be general for the regulation of membrane remodeling. Our work suggests that interplay between membrane lipids and membrane interacting proteins facilitates spatial and temporal coordination of membrane remodeling.
Organizational Affiliation:
The Hormel Institute, University of Minnesota, 801 16(th) Avenue NE, Austin, MN 55912, USA.