Structural insights into the promutagenic bypass of the major cisplatin-induced DNA lesion.
Ouzon-Shubeita, H., Vilas, C.K., Lee, S.(2020) Biochem J 477: 937-951
- PubMed: 32039434
- DOI: https://doi.org/10.1042/BCJ20190906
- Primary Citation of Related Structures:
6U6B - PubMed Abstract:
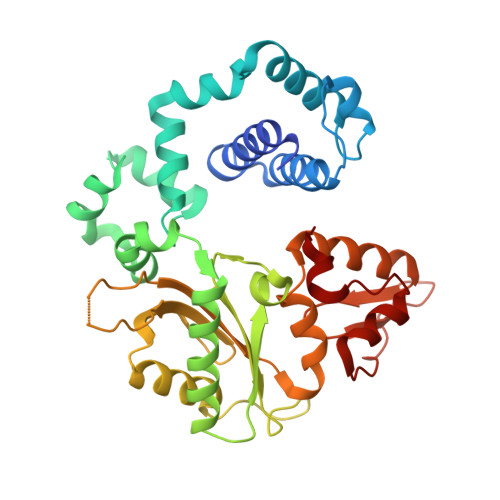
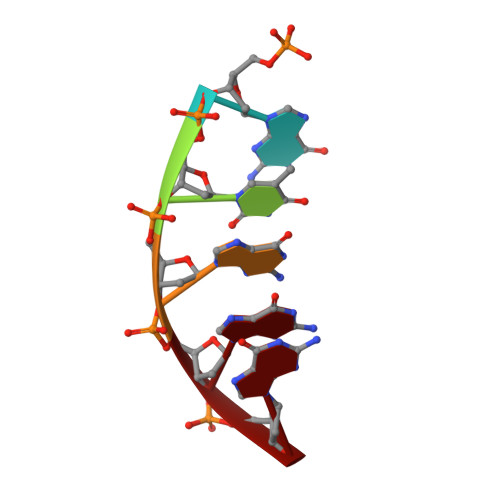
The cisplatin-1,2-d(GpG) (Pt-GG) intrastrand cross-link is the predominant DNA lesion generated by cisplatin. Cisplatin has been shown to predominantly induce G to T mutations and Pt-GG permits significant misincorporation of dATP by human DNA polymerase β (polβ). In agreement, polβ overexpression, which is frequently observed in cancer cells, is linked to cisplatin resistance and a mutator phenotype. However, the structural basis for the misincorporation of dATP opposite Pt-GG is unknown. Here, we report the first structures of a DNA polymerase inaccurately bypassing Pt-GG. We solved two structures of polβ misincorporating dATP opposite the 5'-dG of Pt-GG in the presence of Mg2+ or Mn2+. The Mg2+-bound structure exhibits a sub-optimal conformation for catalysis, while the Mn2+-bound structure is in a catalytically more favorable semi-closed conformation. In both structures, dATP does not form a coplanar base pairing with Pt-GG. In the polβ active site, the syn-dATP opposite Pt-GG appears to be stabilized by protein templating and pi stacking interactions, which resembles the polβ-mediated dATP incorporation opposite an abasic site. Overall, our results suggest that the templating Pt-GG in the polβ active site behaves like an abasic site, promoting the insertion of dATP in a non-instructional manner.
Organizational Affiliation:
Division of Chemical Biology and Medicinal Chemistry, College of Pharmacy, The University of Texas at Austin, Austin, TX 78712, U.S.A.