Molecular Basis for Synaptotagmin-1-Associated Neurodevelopmental Disorder.
Bradberry, M.M., Courtney, N.A., Dominguez, M.J., Lofquist, S.M., Knox, A.T., Sutton, R.B., Chapman, E.R.(2020) Neuron 107: 52-64.e7
- PubMed: 32362337
- DOI: https://doi.org/10.1016/j.neuron.2020.04.003
- Primary Citation of Related Structures:
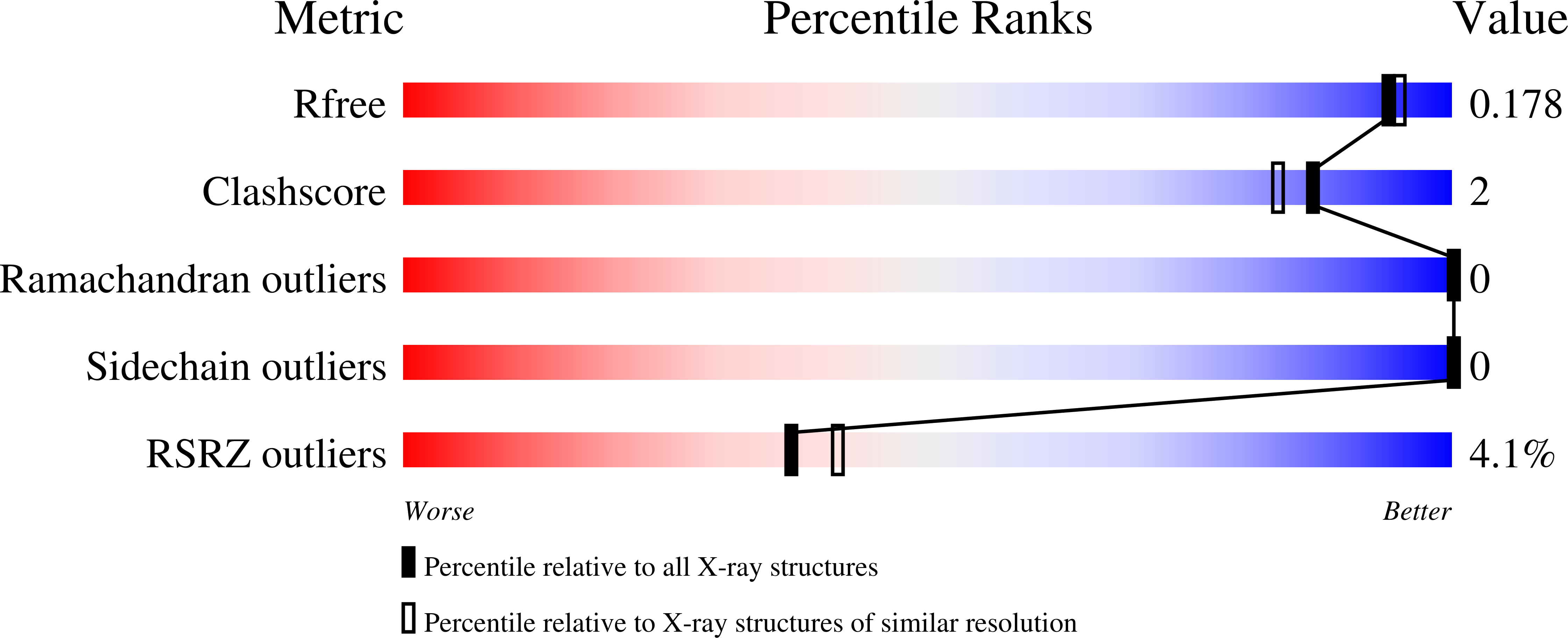
6TZ3, 6U41, 6U4U, 6U4W - PubMed Abstract:
At neuronal synapses, synaptotagmin-1 (syt1) acts as a Ca 2+ sensor that synchronizes neurotransmitter release with Ca 2+ influx during action potential firing. Heterozygous missense mutations in syt1 have recently been associated with a severe but heterogeneous developmental syndrome, termed syt1-associated neurodevelopmental disorder. Well-defined pathogenic mechanisms, and the basis for phenotypic heterogeneity in this disorder, remain unknown. Here, we report the clinical, physiological, and biophysical characterization of three syt1 mutations from human patients. Synaptic transmission was impaired in neurons expressing mutant variants, which demonstrated potent, graded dominant-negative effects. Biophysical interrogation of the mutant variants revealed novel mechanistic features concerning the cooperative action, and functional specialization, of the tandem Ca 2+ -sensing domains of syt1. These mechanistic studies led to the discovery that a clinically approved K + channel antagonist is able to rescue the dominant-negative heterozygous phenotype. Our results establish a molecular cause, basis for phenotypic heterogeneity, and potential treatment approach for syt1-associated neurodevelopmental disorder.
Organizational Affiliation:
Howard Hughes Medical Institute and Department of Neuroscience, University of Wisconsin School of Medicine and Public Health, Madison, WI 53705, USA; Medical Scientist Training Program, University of Wisconsin School of Medicine and Public Health, Madison, WI 53705, USA.