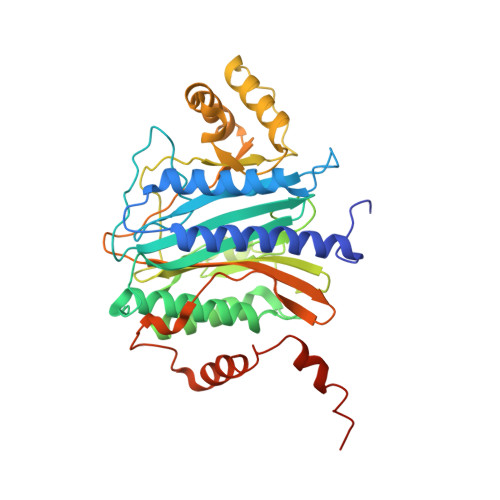
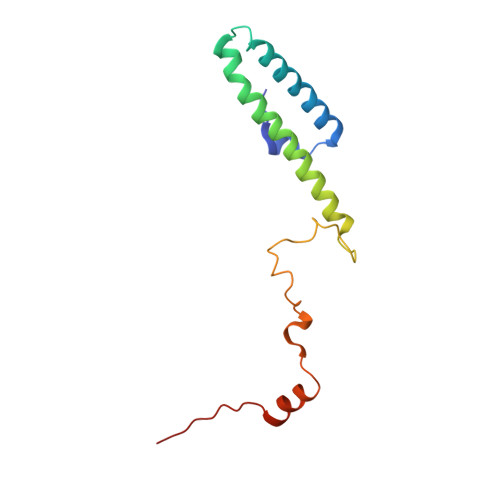
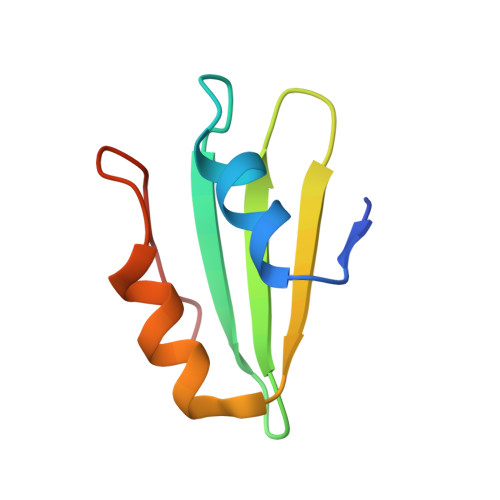
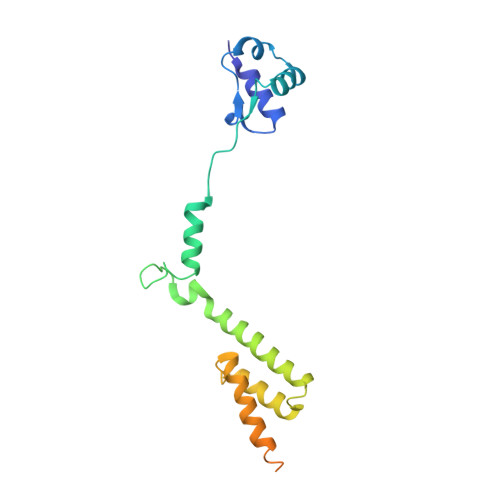
MetAP-like Ebp1 occupies the human ribosomal tunnel exit and recruits flexible rRNA expansion segments.
Wild, K., Aleksic, M., Lapouge, K., Juaire, K.D., Flemming, D., Pfeffer, S., Sinning, I.(2020) Nat Commun 11: 776-776
- PubMed: 32034140
- DOI: https://doi.org/10.1038/s41467-020-14603-7
- Primary Citation of Related Structures:
6SXO - PubMed Abstract:
Human Ebp1 is a member of the proliferation-associated 2G4 (PA2G4) family and plays an important role in cancer regulation. Ebp1 shares the methionine aminopeptidase (MetAP) fold and binds to mature 80S ribosomes for translational control. Here, we present a cryo-EM single particle analysis reconstruction of Ebp1 bound to non-translating human 80S ribosomes at a resolution range from 3.3 to ~8 Å. Ebp1 blocks the tunnel exit with major interactions to the general uL23/uL29 docking site for nascent chain-associated factors complemented by eukaryote-specific eL19 and rRNA helix H59. H59 is defined as dynamic adaptor undergoing significant remodeling upon Ebp1 binding. Ebp1 recruits rRNA expansion segment ES27L to the tunnel exit via specific interactions with rRNA consensus sequences. The Ebp1-ribosome complex serves as a template for MetAP binding and provides insights into the structural principles for spatial coordination of co-translational events and molecular triage at the ribosomal tunnel exit.
Organizational Affiliation:
Biochemiezentrum der Universität Heidelberg (BZH), INF 328, D-69120, Heidelberg, Germany.