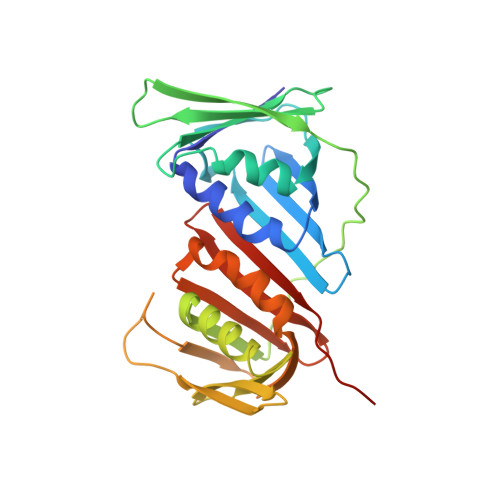
Direct binding of Cdt2 to PCNA is important for targeting the CRL4Cdt2E3 ligase activity to Cdt1.
Hayashi, A., Giakoumakis, N.N., Heidebrecht, T., Ishii, T., Panagopoulos, A., Caillat, C., Takahara, M., Hibbert, R.G., Suenaga, N., Stadnik-Spiewak, M., Takahashi, T., Shiomi, Y., Taraviras, S., von Castelmur, E., Lygerou, Z., Perrakis, A., Nishitani, H.(2018) Life Sci Alliance 1: e201800238-e201800238
- PubMed: 30623174
- DOI: https://doi.org/10.26508/lsa.201800238
- Primary Citation of Related Structures:
6QC0, 6QCG - PubMed Abstract:
The CRL4 Cdt2 ubiquitin ligase complex is an essential regulator of cell-cycle progression and genome stability, ubiquitinating substrates such as p21, Set8, and Cdt1, via a display of substrate degrons on proliferating cell nuclear antigens (PCNAs). Here, we examine the hierarchy of the ligase and substrate recruitment kinetics onto PCNA at sites of DNA replication. We demonstrate that the C-terminal end of Cdt2 bears a PCNA interaction protein motif (PIP box, Cdt2 PIP ), which is necessary and sufficient for the binding of Cdt2 to PCNA. Cdt2 PIP binds PCNA directly with high affinity, two orders of magnitude tighter than the PIP box of Cdt1. X-ray crystallographic structures of PCNA bound to Cdt2 PIP and Cdt1 PIP show that the peptides occupy all three binding sites of the trimeric PCNA ring. Mutating Cdt2 PIP weakens the interaction with PCNA, rendering CRL4 Cdt2 less effective in Cdt1 ubiquitination and leading to defects in Cdt1 degradation. The molecular mechanism we present suggests a new paradigm for bringing substrates to the CRL4-type ligase, where the substrate receptor and substrates bind to a common multivalent docking platform to enable subsequent ubiquitination.
Organizational Affiliation:
Graduate School of Life Science, University of Hyogo, Kamigori, Japan.