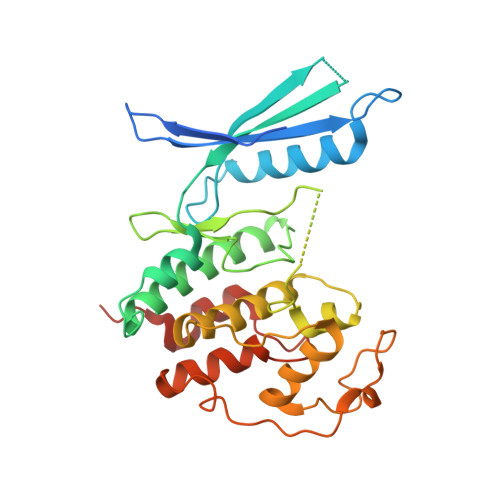
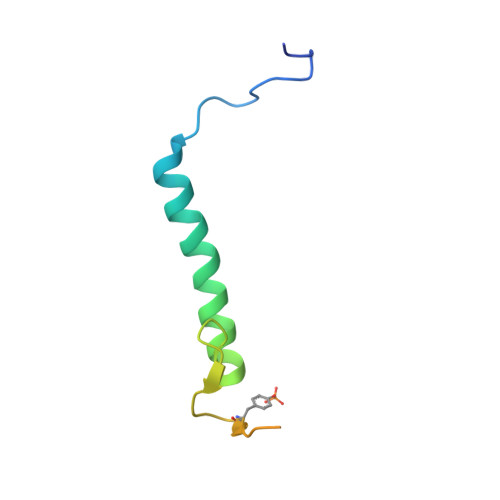
p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Guiley, K.Z., Stevenson, J.W., Lou, K., Barkovich, K.J., Kumarasamy, V., Wijeratne, T.U., Bunch, K.L., Tripathi, S., Knudsen, E.S., Witkiewicz, A.K., Shokat, K.M., Rubin, S.M.(2019) Science 366
- PubMed: 31831640
- DOI: https://doi.org/10.1126/science.aaw2106
- Primary Citation of Related Structures:
6P8E, 6P8F, 6P8G, 6P8H - PubMed Abstract:
The p27 protein is a canonical negative regulator of cell proliferation and acts primarily by inhibiting cyclin-dependent kinases (CDKs). Under some circumstances, p27 is associated with active CDK4, but no mechanism for activation has been described. We found that p27, when phosphorylated by tyrosine kinases, allosterically activated CDK4 in complex with cyclin D1 (CDK4-CycD1). Structural and biochemical data revealed that binding of phosphorylated p27 (phosp27) to CDK4 altered the kinase adenosine triphosphate site to promote phosphorylation of the retinoblastoma tumor suppressor protein (Rb) and other substrates. Surprisingly, purified and endogenous phosp27-CDK4-CycD1 complexes were insensitive to the CDK4-targeting drug palbociclib. Palbociclib instead primarily targeted monomeric CDK4 and CDK6 (CDK4/6) in breast tumor cells. Our data characterize phosp27-CDK4-CycD1 as an active Rb kinase that is refractory to clinically relevant CDK4/6 inhibitors.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, Santa Cruz, CA 95064, USA.