COA6 Is Structurally Tuned to Function as a Thiol-Disulfide Oxidoreductase in Copper Delivery to Mitochondrial Cytochrome c Oxidase.
Soma, S., Morgada, M.N., Naik, M.T., Boulet, A., Roesler, A.A., Dziuba, N., Ghosh, A., Yu, Q., Lindahl, P.A., Ames, J.B., Leary, S.C., Vila, A.J., Gohil, V.M.(2019) Cell Rep 29: 4114-4126.e5
- PubMed: 31851937
- DOI: https://doi.org/10.1016/j.celrep.2019.11.054
- Primary Citation of Related Structures:
6NL3 - PubMed Abstract:
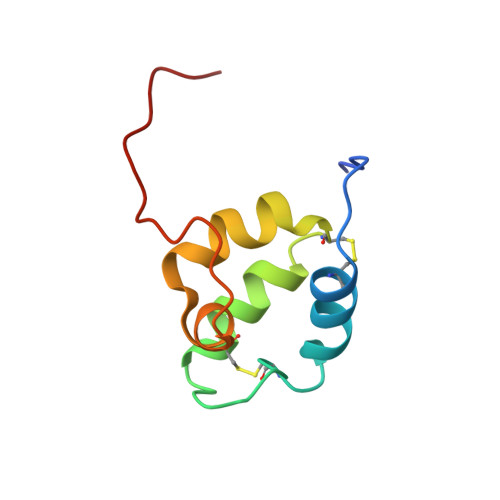
In eukaryotes, cellular respiration is driven by mitochondrial cytochrome c oxidase (CcO), an enzyme complex that requires copper cofactors for its catalytic activity. Insertion of copper into its catalytically active subunits, including COX2, is a complex process that requires metallochaperones and redox proteins including SCO1, SCO2, and COA6, a recently discovered protein whose molecular function is unknown. To uncover the molecular mechanism by which COA6 and SCO proteins mediate copper delivery to COX2, we have solved the solution structure of COA6, which reveals a coiled-coil-helix-coiled-coil-helix domain typical of redox-active proteins found in the mitochondrial inter-membrane space. Accordingly, we demonstrate that COA6 can reduce the copper-coordinating disulfides of its client proteins, SCO1 and COX2, allowing for copper binding. Finally, our determination of the interaction surfaces and reduction potentials of COA6 and its client proteins provides a mechanism of how metallochaperone and disulfide reductase activities are coordinated to deliver copper to CcO.
Organizational Affiliation:
Department of Biochemistry and Biophysics, MS 3474, Texas A&M University, College Station, TX 77843, USA.