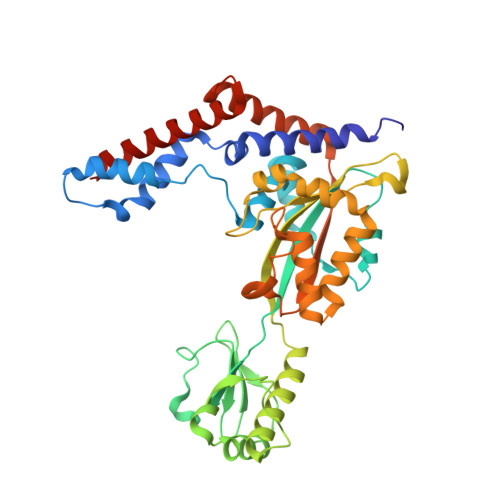
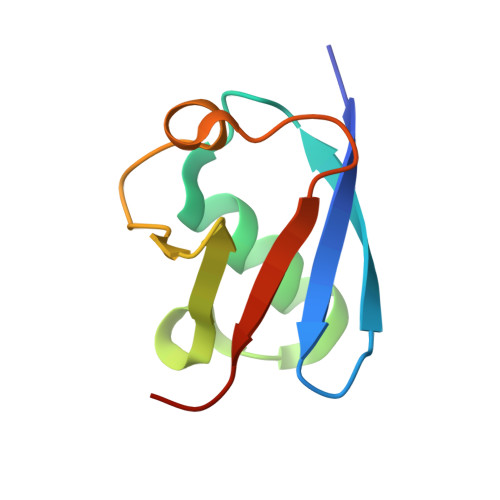
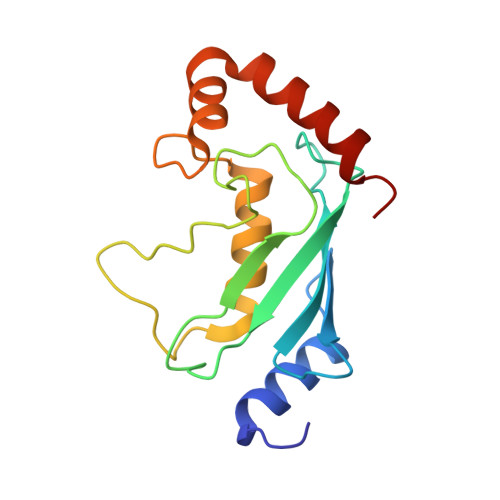
Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC.
Mu, Y., Wang, Y., Huang, Y., Li, D., Han, Y., Chang, M., Fu, J., Xie, Y., Ren, J., Wang, H., Zhang, Y., Luo, Z.Q., Feng, Y.(2020) Nat Commun 11: 1774-1774
- PubMed: 32286321
- DOI: https://doi.org/10.1038/s41467-020-15645-7
- Primary Citation of Related Structures:
6K3B, 6KFP, 6KG6 - PubMed Abstract:
Protein ubiquitination is one of the most prevalent post-translational modifications, controlling virtually every process in eukaryotic cells. Recently, the Legionella effector MavC was found to mediate a unique ubiquitination through transglutamination, linking ubiquitin (Ub) to UBE2N through Ub Gln40 in a process that can be inhibited by another Legionella effector, Lpg2149. Here, we report the structures of MavC/UBE2N/Ub ternary complex, MavC/UBE2N-Ub (product) binary complex, and MavC/Lpg2149 binary complex. During the ubiquitination, the loop containing the modification site K92 of UBE2N undergoes marked conformational change, and Lpg2149 inhibits this ubiquitination through competing with Ub to bind MavC. Moreover, we found that MavC itself also exhibits weak deubiquitinase activity towards this non-canonical ubiquitination. Together, our study not only provides insights into the mechanism and inhibition of this transglutaminase-induced ubiquitination by MavC, but also sheds light on the future studies into UBE2N inhibition by this modification and deubiquitinases of this unique ubiquitination.
Organizational Affiliation:
Beijing Advanced Innovation Center for Soft Matter Science and Engineering, Beijing Key Laboratory of Bioprocess, State Key Laboratory of Chemical Resource Engineering, College of Life Science and Technology, Beijing University of Chemical Technology, 100029, Beijing, China.