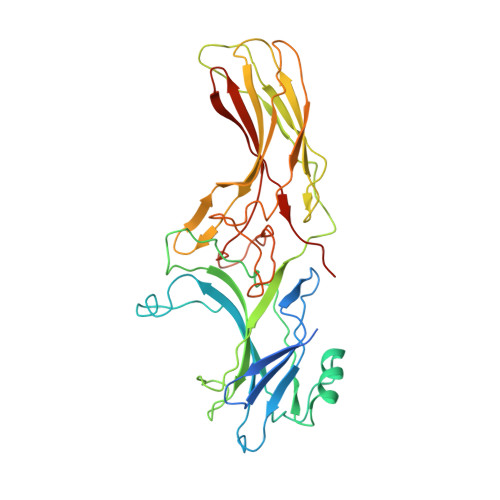
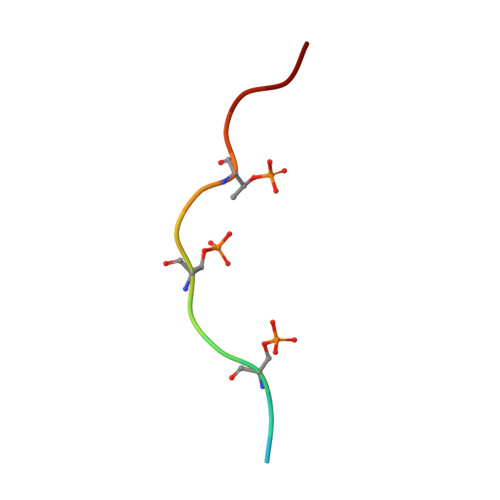
Crystal Structure of beta-Arrestin 2 in Complex with CXCR7 Phosphopeptide.
Min, K., Yoon, H.J., Park, J.Y., Baidya, M., Dwivedi-Agnihotri, H., Maharana, J., Chaturvedi, M., Chung, K.Y., Shukla, A.K., Lee, H.H.(2020) Structure 28: 1014-1023.e4
- PubMed: 32579945
- DOI: https://doi.org/10.1016/j.str.2020.06.002
- Primary Citation of Related Structures:
6K3F - PubMed Abstract:
β-Arrestins (βarrs) critically regulate G-protein-coupled receptor (GPCR) signaling and trafficking. βarrs have two isoforms, βarr1 and βarr2. Receptor phosphorylation is a key determinant for the binding of βarrs, and understanding the intricate details of receptor-βarr interaction is the next frontier in GPCR structural biology. The high-resolution structure of active βarr1 in complex with a phosphopeptide derived from GPCR has been revealed, but that of βarr2 remains elusive. Here, we present a 2.3-Å crystal structure of βarr2 in complex with a phosphopeptide (C7pp) derived from the carboxyl terminus of CXCR7. The structural analysis of C7pp-bound βarr2 reveals key differences from the previously determined active conformation of βarr1. One of the key differences is that C7pp-bound βarr2 shows a relatively small inter-domain rotation. Antibody-fragment-based conformational sensor and hydrogen/deuterium exchange experiments further corroborated the structural features of βarr2 and suggested that βarr2 adopts a range of inter-domain rotations.
Organizational Affiliation:
Department of Chemistry, College of Natural Sciences, Seoul National University, Seoul 08826, Republic of Korea.