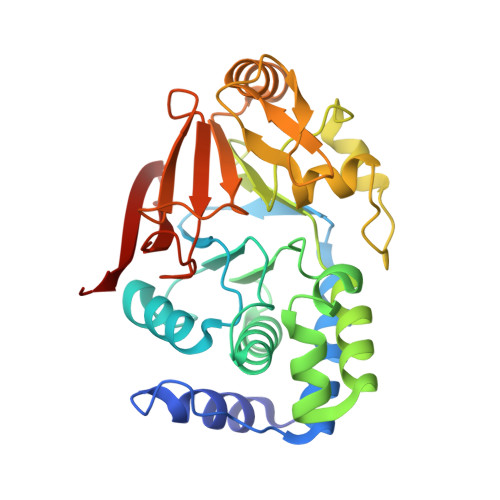
Identification of the substrate recruitment mechanism of the muscle glycogen protein phosphatase 1 holoenzyme.
Kumar, G.S., Choy, M.S., Koveal, D.M., Lorinsky, M.K., Lyons, S.P., Kettenbach, A.N., Page, R., Peti, W.(2018) Sci Adv 4: eaau6044-eaau6044
- PubMed: 30443599
- DOI: https://doi.org/10.1126/sciadv.aau6044
- Primary Citation of Related Structures:
6DNO - PubMed Abstract:
Glycogen is the primary storage form of glucose. Glycogen synthesis and breakdown are tightly controlled by glycogen synthase (GYS) and phosphorylase, respectively. The enzyme responsible for dephosphorylating GYS and phosphorylase, which results in their activation (GYS) or inactivation (phosphorylase) to robustly stimulate glycogen synthesis, is protein phosphatase 1 (PP1). However, our understanding of how PP1 recruits these substrates is limited. Here, we show how PP1, together with its muscle glycogen-targeting (G M ) regulatory subunit, recruits and selectively dephosphorylates its substrates. Our molecular data reveal that the G M carbohydrate binding module (G M CBM21 ), which is amino-terminal to the G M PP1 binding domain, has a dual function in directing PP1 substrate specificity: It either directly recruits substrates (i.e., GYS) or recruits them indirectly by localization (via glycogen for phosphorylase). Our data provide the molecular basis for PP1 regulation by G M and reveal how PP1-mediated dephosphorylation is driven by scaffolding-based substrate recruitment.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of Arizona, Tucson, AZ 85721, USA.