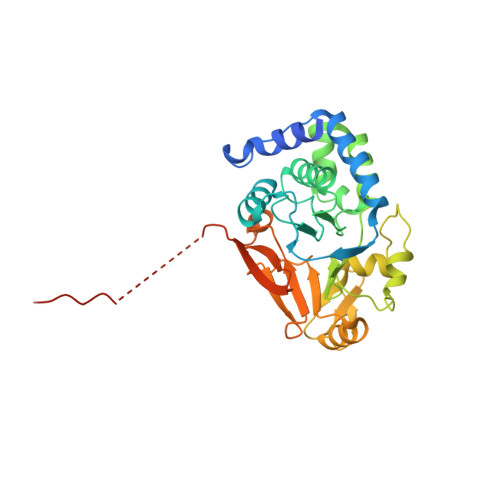
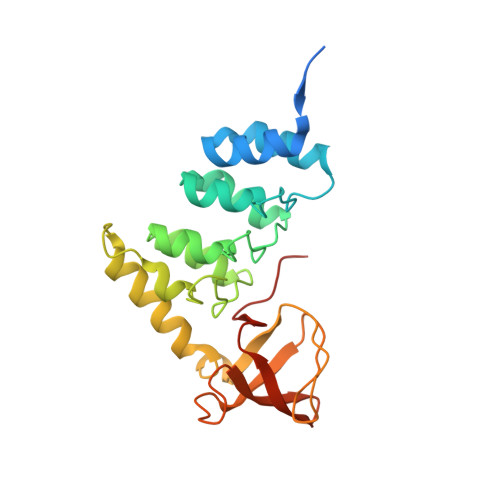
Flexible Tethering of ASPP Proteins Facilitates PP-1c Catalysis.
Zhou, Y., Millott, R., Kim, H.J., Peng, S., Edwards, R.A., Skene-Arnold, T., Hammel, M., Lees-Miller, S.P., Tainer, J.A., Holmes, C.F.B., Glover, J.N.M.(2019) Structure 27: 1485-1496.e4
- PubMed: 31402222
- DOI: https://doi.org/10.1016/j.str.2019.07.012
- Primary Citation of Related Structures:
6DCX - PubMed Abstract:
ASPP (apoptosis-stimulating proteins of p53) proteins bind PP-1c (protein phosphatase 1) and regulate p53 impacting cancer cell growth and apoptosis. Here we determine the crystal structure of the oncogenic ASPP protein, iASPP, bound to PP-1c. The structure reveals a 1:1 complex that relies on interactions of the iASPP SILK and RVxF motifs with PP-1c, plus interactions of the PP-1c PxxPxR motif with the iASPP SH3 domain. Small-angle X-ray scattering analyses suggest that the crystal structure undergoes slow interconversion with more extended conformations in solution. We show that iASPP, and the tumor suppressor ASPP2, enhance the catalytic activity of PP-1c against the small-molecule substrate, pNPP as well as p53. The combined results suggest that PxxPxR binding to iASPP SH3 domain is critical for complex formation, and that the modular ASPP-PP-1c interface provides dynamic flexibility that enables functional binding and dephosphorylation of p53 and other diverse protein substrates.
Organizational Affiliation:
Department of Biochemistry, University of Alberta, Edmonton, AB T6G 2H7, Canada.