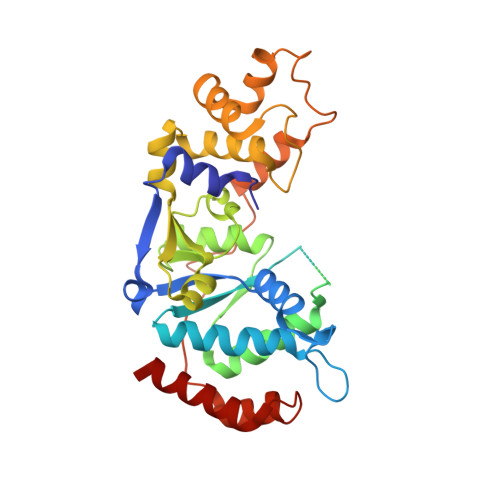
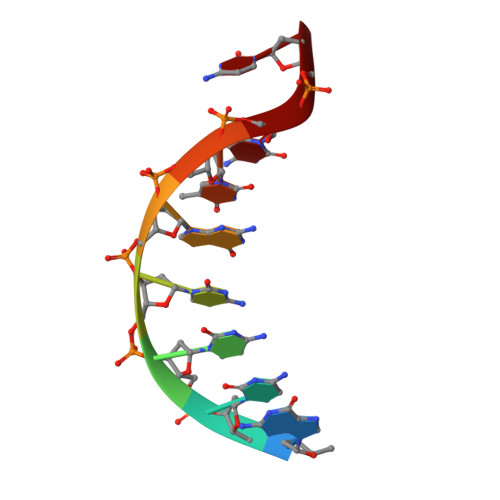
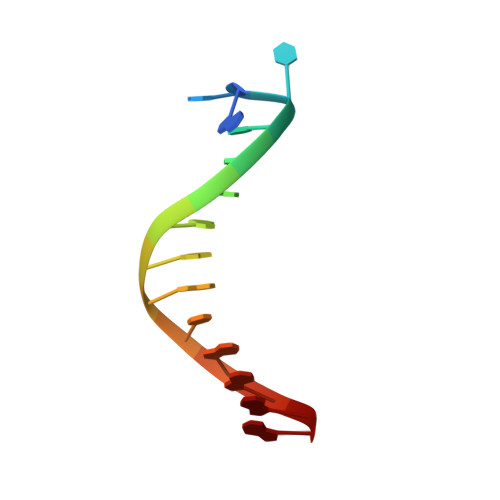
Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Xu, H., Shi, R., Han, W., Cheng, J., Xu, X., Cheng, K., Wang, L., Tian, B., Zheng, L., Shen, B., Hua, Y., Zhao, Y.(2018) Nucleic Acids Res 46: 11315-11325
- PubMed: 30295841
- DOI: https://doi.org/10.1093/nar/gky911
- Primary Citation of Related Structures:
5ZOD, 5ZOE, 5ZOF, 5ZOG - PubMed Abstract:
Human flap endonuclease 1 (hFEN1) is a structure-specific nuclease essential for DNA replication and repair processes. hFEN1 has 5' flap removal activity, as well as gap endonuclease activity that is critical for restarting stalled replication forks. Here, we report the crystal structures of wild-type and mutant hFEN1 proteins in complex with DNA substrates, followed by mutagenesis studies that provide mechanistic insight into the protein-protein interactions of hFEN1. We found that in an α-helix forming the helical gateway of hFEN1 recognizes the 5' flap prior to its threading into the active site for cleavage. We also found that the β-pin region is rigidified into a short helix in R192F hFEN1-DNA structures, suppressing its gap endonuclease activity and cycle-dependent kinase interactions. Our findings suggest that a single mutation at the primary methylation site can alter the function of hFEN1 and provide insight into the role of the β-pin region in hFEN1 protein interactions that are essential for DNA replication and repair.
Organizational Affiliation:
Key Laboratory of Chinese Ministry of Agriculture for Nuclear-Agricultural Sciences, Institute of Nuclear-Agricultural Sciences, Zhejiang University, Hangzhou, Zhejiang 310029, China.