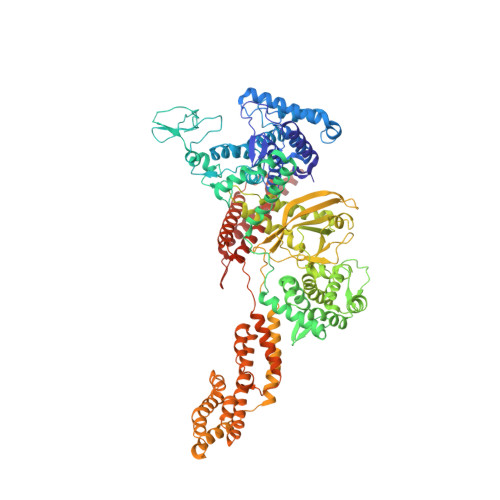
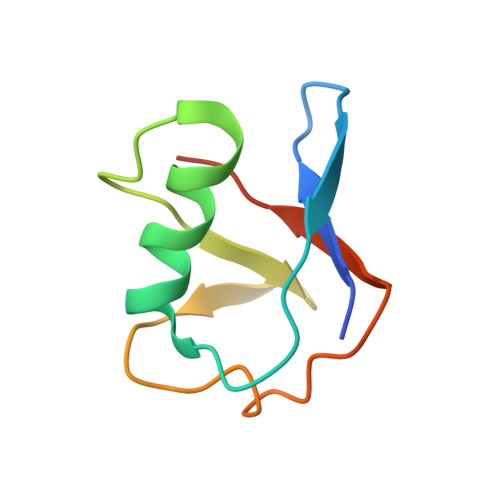
Structural basis of ubiquitin modification by the Legionella effector SdeA.
Dong, Y., Mu, Y., Xie, Y., Zhang, Y., Han, Y., Zhou, Y., Wang, W., Liu, Z., Wu, M., Wang, H., Pan, M., Xu, N., Xu, C.Q., Yang, M., Fan, S., Deng, H., Tan, T., Liu, X., Liu, L., Li, J., Wang, J., Fang, X., Feng, Y.(2018) Nature 557: 674-678
- PubMed: 29795342
- DOI: https://doi.org/10.1038/s41586-018-0146-7
- Primary Citation of Related Structures:
5YIJ, 5YIK, 5YIM - PubMed Abstract:
Protein ubiquitination is a multifaceted post-translational modification that controls almost every process in eukaryotic cells. Recently, the Legionella effector SdeA was reported to mediate a unique phosphoribosyl-linked ubiquitination through successive modifications of the Arg42 of ubiquitin (Ub) by its mono-ADP-ribosyltransferase (mART) and phosphodiesterase (PDE) domains. However, the mechanisms of SdeA-mediated Ub modification and phosphoribosyl-linked ubiquitination remain unknown. Here we report the structures of SdeA in its ligand-free, Ub-bound and Ub-NADH-bound states. The structures reveal that the mART and PDE domains of SdeA form a catalytic domain over its C-terminal region. Upon Ub binding, the canonical ADP-ribosyltransferase toxin turn-turn (ARTT) and phosphate-nicotinamide (PN) loops in the mART domain of SdeA undergo marked conformational changes. The Ub Arg72 might act as a 'probe' that interacts with the mART domain first, and then movements may occur in the side chains of Arg72 and Arg42 during the ADP-ribosylation of Ub. Our study reveals the mechanism of SdeA-mediated Ub modification and provides a framework for further investigations into the phosphoribosyl-linked ubiquitination process.
Organizational Affiliation:
Beijing Advanced Innovation Center for Soft Matter Science and Engineering, Beijing Key Laboratory of Bioprocess, College of Life Science and Technology, Beijing University of Chemical Technology, Beijing, China.