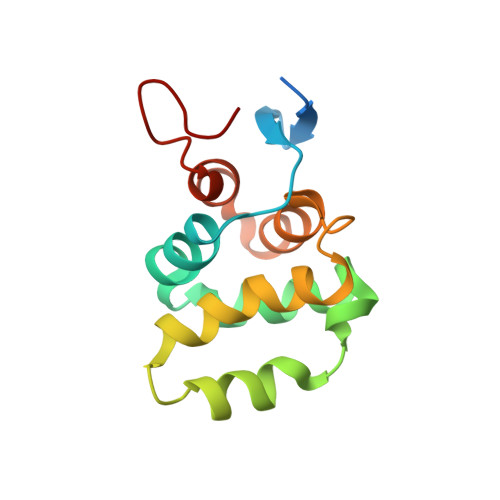
Structure of the C-terminal domain of TRADD reveals a novel fold in the death domain superfamily.
Zhang, N., Yuan, W., Fan, J.S., Lin, Z.(2017) Sci Rep 7: 7073-7073
- PubMed: 28765645
- DOI: https://doi.org/10.1038/s41598-017-07348-9
- Primary Citation of Related Structures:
5XME - PubMed Abstract:
The TNFR1-associated death domain protein (TRADD) is an intracellular adaptor protein involved in various signaling pathways, such as antiapoptosis. Its C-terminal death domain (DD) is responsible for binding other DD-containing proteins including the p75 neurotrophin receptor (p75 NTR ). Here we present a solution structure of TRADD DD derived from high-resolution NMR spectroscopy. The TRADD DD comprises two super-secondary structures, an all-helix Greek key motif and a β-hairpin motif flanked by two α helices, which make it unique among all known DD structures. The β-hairpin motif is essential for TRADD DD to fold into a functional globular domain. The highly-charged surface suggests a critical role of electrostatic interactions in TRADD DD-mediated signaling. This novel structure represents a new class within the DD superfamily and provides a structural basis for studying homotypic DD interactions. NMR titration revealed a direct weak interaction between TRADD DD and p75 NTR DD monomers. A binding site next to the p75 NTR DD homodimerization interface indicates that TRADD DD recruitment to p75 NTR requires separation of the p75 NTR DD homodimer, explaining the mechanism of NGF-dependent activation of p75 NTR -TRADD-mediated antiapoptotic pathway in breast cancer cell.
Organizational Affiliation:
School of Life Sciences, Tianjin University, Tianjin, 300072, P.R. China.