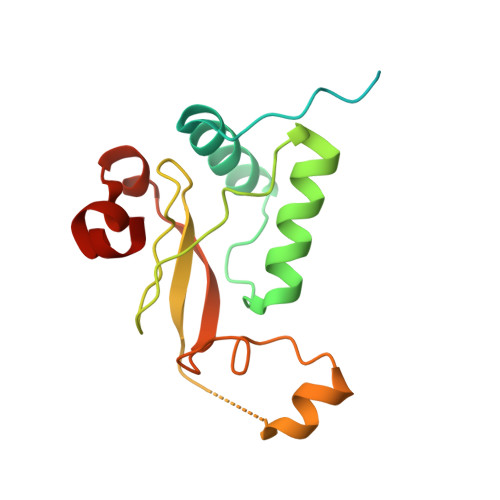
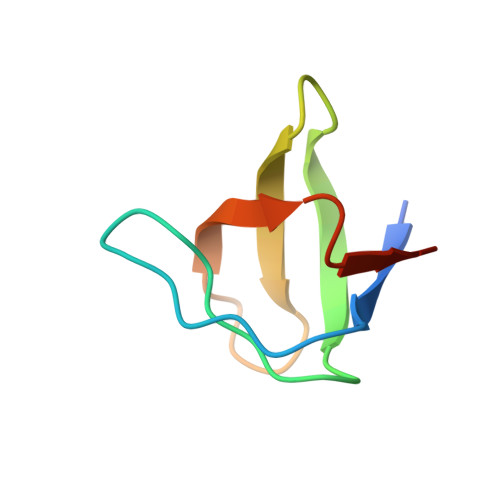
Endocytic sorting motif interactions involved in Nef-mediated downmodulation of CD4 and CD3.
Manrique, S., Sauter, D., Horenkamp, F.A., Lulf, S., Yu, H., Hotter, D., Anand, K., Kirchhoff, F., Geyer, M.(2017) Nat Commun 8: 442-442
- PubMed: 28874665
- DOI: https://doi.org/10.1038/s41467-017-00481-z
- Primary Citation of Related Structures:
5NUH, 5NUI - PubMed Abstract:
Lentiviral Nefs recruit assembly polypeptide complexes and target sorting motifs in cellular receptors to induce their internalization. While Nef-mediated CD4 downmodulation is conserved, the ability to internalize CD3 was lost in HIV-1 and its precursors. Although both functions play key roles in lentiviral replication and pathogenicity, the underlying structural requirements are poorly defined. Here, we determine the structure of SIV mac239 Nef bound to the ExxxLM motif of another Nef molecule at 2.5 Å resolution. This provides a basis for a structural model, where a hydrophobic crevice in simian immunodeficiency virus (SIV) Nef targets a dileucine motif in CD4 and a tyrosine-based motif in CD3. Introducing key residues into this crevice of HIV-1 Nef enables CD3 binding but an additional N-terminal tyrosine motif is required for internalization. Our resolution of the CD4/Nef/AP2 complex and generation of HIV-1 Nefs capable of CD3 downregulation provide insights into sorting motif interactions and target discrimination of Nef.HIV and simian immunodeficiency virus (SIV) Nef proteins both stimulate the clathrin-mediated endocytosis of CD4 but differ in downmodulation of the immune receptor CD3. Here, the authors present the structure of SIV Nef bound to the ExxxLM motif of another Nef molecule, which allows them to propose a model how Nef recognizes these motifs in CD3 and CD4.
Organizational Affiliation:
Institute of Innate Immunity, Department of Structural Immunology, University of Bonn, Sigmund-Freud-Str. 25, 53127, Bonn, Germany.