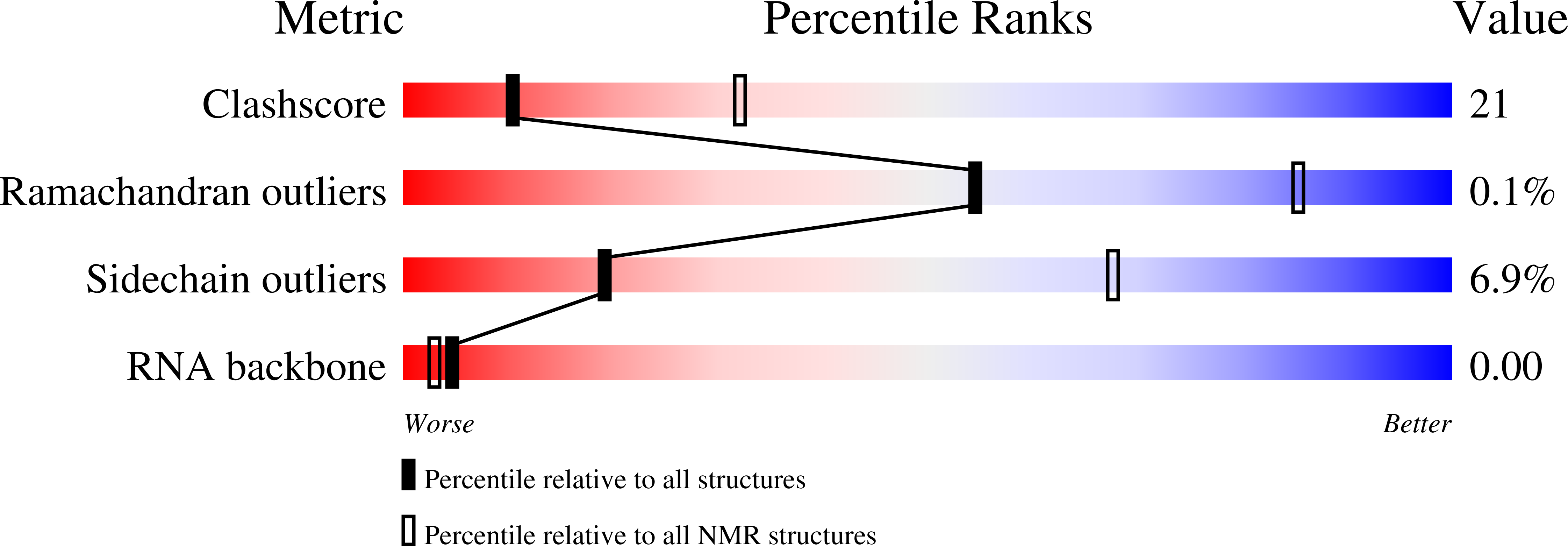
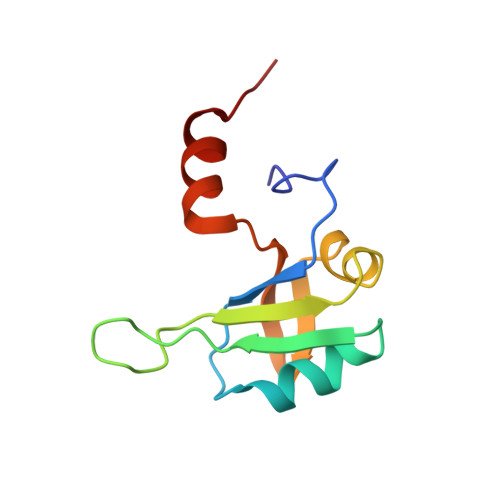
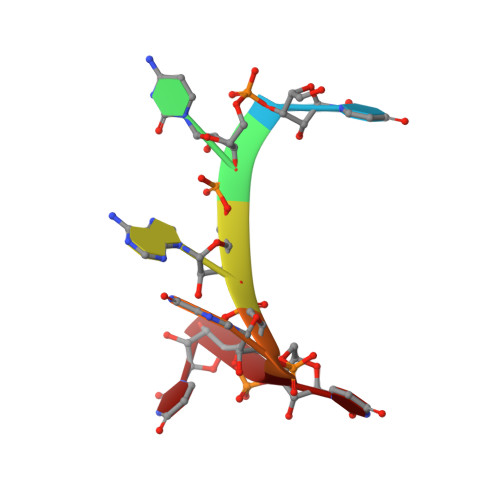
Tandem hnRNP A1 RNA recognition motifs act in concert to repress the splicing of survival motor neuron exon 7.
Beusch, I., Barraud, P., Moursy, A., Clery, A., Allain, F.H.(2017) Elife 6
- PubMed: 28650318
- DOI: https://doi.org/10.7554/eLife.25736
- Primary Citation of Related Structures:
5MPG, 5MPL - PubMed Abstract:
HnRNP A1 regulates many alternative splicing events by the recognition of splicing silencer elements. Here, we provide the solution structures of its two RNA recognition motifs (RRMs) in complex with short RNA. In addition, we show by NMR that both RRMs of hnRNP A1 can bind simultaneously to a single bipartite motif of the human intronic splicing silencer ISS-N1, which controls survival of motor neuron exon 7 splicing. RRM2 binds to the upstream motif and RRM1 to the downstream motif. Combining the insights from the structure with in cell splicing assays we show that the architecture and organization of the two RRMs is essential to hnRNP A1 function. The disruption of the inter-RRM interaction or the loss of RNA binding capacity of either RRM impairs splicing repression by hnRNP A1. Furthermore, both binding sites within the ISS-N1 are important for splicing repression and their contributions are cumulative rather than synergistic.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, Eidgenössische Technische Hochschule Zürich, Zürich, Switzerland.