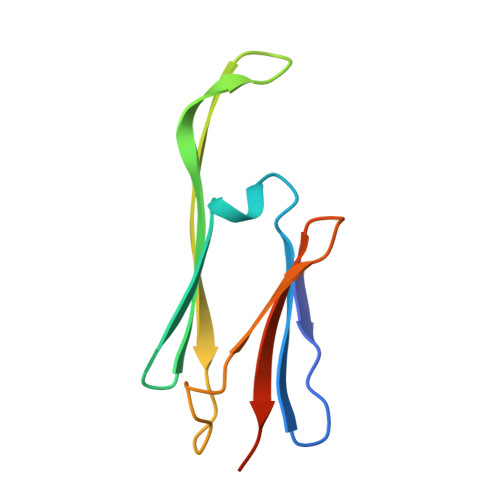
The structured core domain of alpha B-crystallin can prevent amyloid fibrillation and associated toxicity.
Hochberg, G.K., Ecroyd, H., Liu, C., Cox, D., Cascio, D., Sawaya, M.R., Collier, M.P., Stroud, J., Carver, J.A., Baldwin, A.J., Robinson, C.V., Eisenberg, D.S., Benesch, J.L., Laganowsky, A.(2014) Proc Natl Acad Sci U S A 111: E1562-E1570
- PubMed: 24711386
- DOI: https://doi.org/10.1073/pnas.1322673111
- Primary Citation of Related Structures:
4M5S, 4M5T, 4MJH - PubMed Abstract:
Mammalian small heat-shock proteins (sHSPs) are molecular chaperones that form polydisperse and dynamic complexes with target proteins, serving as a first line of defense in preventing their aggregation into either amorphous deposits or amyloid fibrils. Their apparently broad target specificity makes sHSPs attractive for investigating ways to tackle disorders of protein aggregation. The two most abundant sHSPs in human tissue are αB-crystallin (ABC) and HSP27; here we present high-resolution structures of their core domains (cABC, cHSP27), each in complex with a segment of their respective C-terminal regions. We find that both truncated proteins dimerize, and although this interface is labile in the case of cABC, in cHSP27 the dimer can be cross-linked by an intermonomer disulfide linkage. Using cHSP27 as a template, we have designed an equivalently locked cABC to enable us to investigate the functional role played by oligomerization, disordered N and C termini, subunit exchange, and variable dimer interfaces in ABC. We have assayed the ability of the different forms of ABC to prevent protein aggregation in vitro. Remarkably, we find that cABC has chaperone activity comparable to that of the full-length protein, even when monomer dissociation is restricted through disulfide linkage. Furthermore, cABC is a potent inhibitor of amyloid fibril formation and, by slowing the rate of its aggregation, effectively reduces the toxicity of amyloid-β peptide to cells. Overall we present a small chaperone unit together with its atomic coordinates that potentially enables the rational design of more effective chaperones and amyloid inhibitors.
Organizational Affiliation:
Physical and Theoretical Chemistry Laboratory, Department of Chemistry, University of Oxford, Oxford OX1 3QZ, United Kingdom.