The Structural Motifs for Substrate Binding and Dimerization of the Alpha Subunit of Collagen Prolyl 4-Hydroxylase
Anantharajan, J., Koski, M.K., Kursula, P., Hieta, R., Bergmann, U., Myllyharju, J., Wierenga, R.K.(2013) Structure 21: 2107
- PubMed: 24207127
- DOI: https://doi.org/10.1016/j.str.2013.09.005
- Primary Citation of Related Structures:
2YQ8, 4BT8, 4BT9, 4BTA, 4BTB - PubMed Abstract:
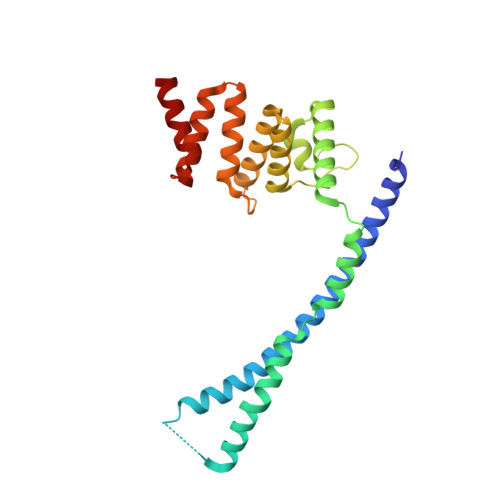
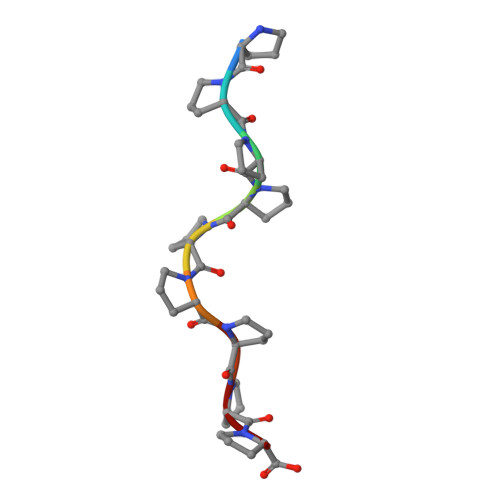
Collagen prolyl 4-hydroxylase (C-P4H) catalyzes the proline hydroxylation of procollagen, an essential modification in the maturation of collagens. C-P4H consists of two catalytic α subunits and two protein disulfide isomerase β subunits. The assembly of these subunits is unknown. The α subunit contains an N domain (1-143), a peptide-substrate-binding-domain (PSB, 144-244) and a catalytic domain (245-517). Here, we report the dimeric structure of the N-terminal region (1-244) of the α subunit. It is shown that the N domain has an important role in the assembly of the C-P4H tetramer, by forming an extended four-helix bundle that includes an antiparallel coiled-coil dimerization motif between the two α subunits. Complexes of this construct with a C-P4H inhibitor and substrate show the mode of peptide-binding to the PSB domain. Both peptides adopt a poly-(L)-proline-type-II helix conformation and bind in a curved, asymmetric groove lined by conserved tyrosines and an Arg-Asp salt bridge.
Organizational Affiliation:
Biocenter Oulu and Department of Biochemistry, University of Oulu, P.O. Box 3000, FIN-90014 Oulu, Finland.