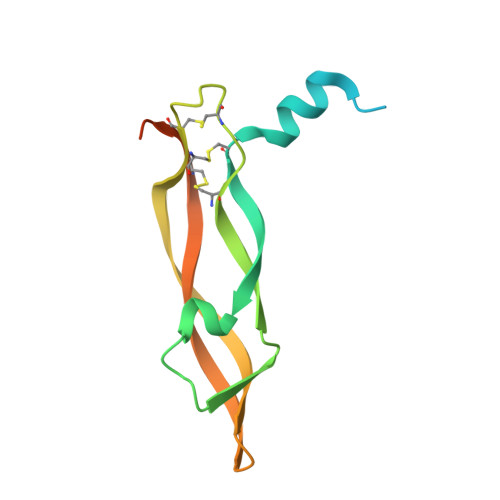
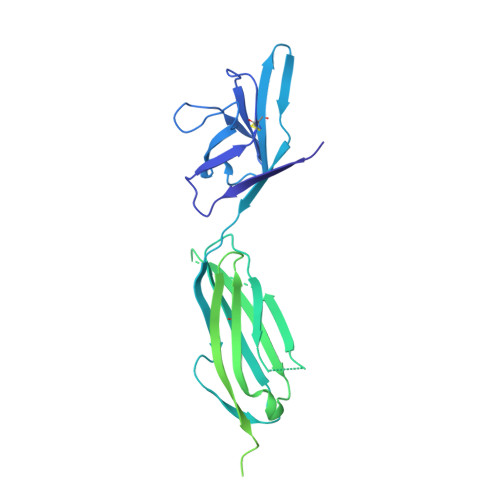
Thermodynamic and structural description of allosterically regulated VEGFR-2 dimerization.
Brozzo, M.S., Bjelic, S., Kisko, K., Schleier, T., Leppanen, V.M., Alitalo, K., Winkler, F.K., Ballmer-Hofer, K.(2012) Blood 119: 1781-1788
- PubMed: 22207738
- DOI: https://doi.org/10.1182/blood-2011-11-390922
- Primary Citation of Related Structures:
3V2A, 3V6B - PubMed Abstract:
VEGFs activate 3 receptor tyrosine kinases, VEGFR-1, VEGFR-2, and VEGFR-3, promoting angiogenic and lymphangiogenic signaling. The extracellular receptor domain (ECD) consists of 7 Ig-homology domains; domains 2 and 3 (D23) represent the ligand-binding domain, whereas the function of D4-7 is unclear. Ligand binding promotes receptor dimerization and instigates transmembrane signaling and receptor kinase activation. In the present study, isothermal titration calorimetry showed that the Gibbs free energy of VEGF-A, VEGF-C, or VEGF-E binding to D23 or the full-length ECD of VEGFR-2 is dominated by favorable entropic contribution with enthalpic penalty. The free energy of VEGF binding to the ECD is 1.0-1.7 kcal/mol less favorable than for binding to D23. A model of the VEGF-E/VEGFR-2 ECD complex derived from small-angle scattering data provided evidence for homotypic interactions in D4-7. We also solved the crystal structures of complexes between VEGF-A or VEGF-E with D23, which revealed comparable binding surfaces and similar interactions between the ligands and the receptor, but showed variation in D23 twist angles. The energetically unfavorable homotypic interactions in D4-7 may be required for re-orientation of receptor monomers, and this mechanism might prevent ligand-independent activation of VEGFR-2 to evade the deleterious consequences for blood and lymph vessel homeostasis arising from inappropriate receptor activation.
Organizational Affiliation:
Biomolecular Research, Molecular Cell Biology Paul Scherrer Institut, Villigen, Switzerland.