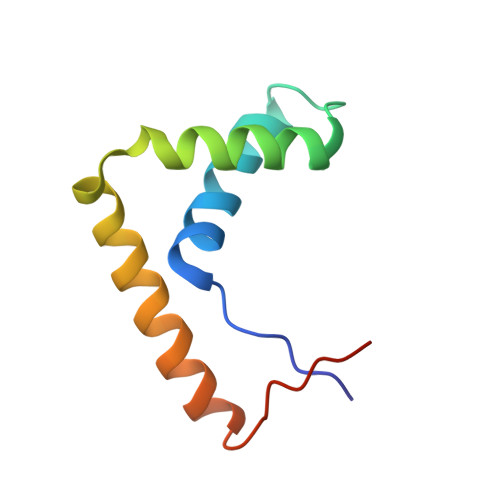
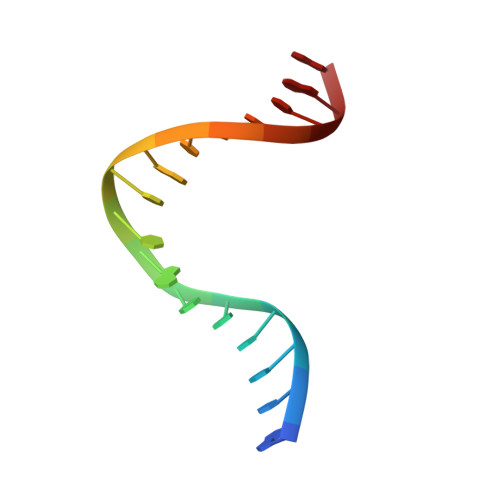
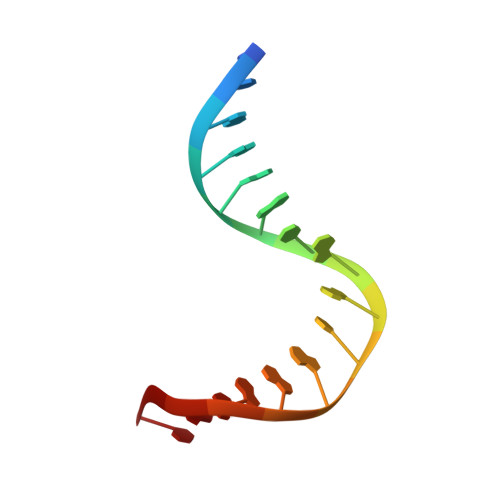
The crystal structure of the Sox4 HMG domain-DNA complex suggests a mechanism for positional interdependence in DNA recognition
Jauch, R., Ng, C.K.L., Narasimhan, K., Kolatkar, P.R.(2012) Biochem J 443: 39-47
- PubMed: 22181698
- DOI: https://doi.org/10.1042/BJ20111768
- Primary Citation of Related Structures:
3U2B - PubMed Abstract:
It has recently been proposed that the sequence preferences of DNA-binding TFs (transcription factors) can be well described by models that include the positional interdependence of the nucleotides of the target sites. Such binding models allow for multiple motifs to be invoked, such as principal and secondary motifs differing at two or more nucleotide positions. However, the structural mechanisms underlying the accommodation of such variant motifs by TFs remain elusive. In the present study we examine the crystal structure of the HMG (high-mobility group) domain of Sox4 [Sry (sex-determining region on the Y chromosome)-related HMG box 4] bound to DNA. By comparing this structure with previously solved structures of Sox17 and Sox2, we observed subtle conformational differences at the DNA-binding interface. Furthermore, using quantitative electrophoretic mobility-shift assays we validated the positional interdependence of two nucleotides and the presence of a secondary Sox motif in the affinity landscape of Sox4. These results suggest that a concerted rearrangement of two interface amino acids enables Sox4 to accommodate primary and secondary motifs. The structural adaptations lead to altered dinucleotide preferences that mutually reinforce each other. These analyses underline the complexity of the DNA recognition by TFs and provide an experimental validation for the conceptual framework of positional interdependence and secondary binding motifs.
Organizational Affiliation:
Laboratory for Structural Biochemistry, Genome Institute of Singapore, 60 Biopolis St, Singapore 138672. jauchr@gis.a-star.edu.sg