The Biological Buffer Bicarbonate/CO(2) Potentiates H(2)O(2)-Mediated Inactivation of Protein Tyrosine Phosphatases.
Zhou, H., Singh, H., Parsons, Z.D., Lewis, S.M., Bhattacharya, S., Seiner, D.R., Labutti, J.N., Reilly, T.J., Tanner, J.J., Gates, K.S.(2011) J Am Chem Soc 133: 15803-15805
- PubMed: 21913686
- DOI: https://doi.org/10.1021/ja2077137
- Primary Citation of Related Structures:
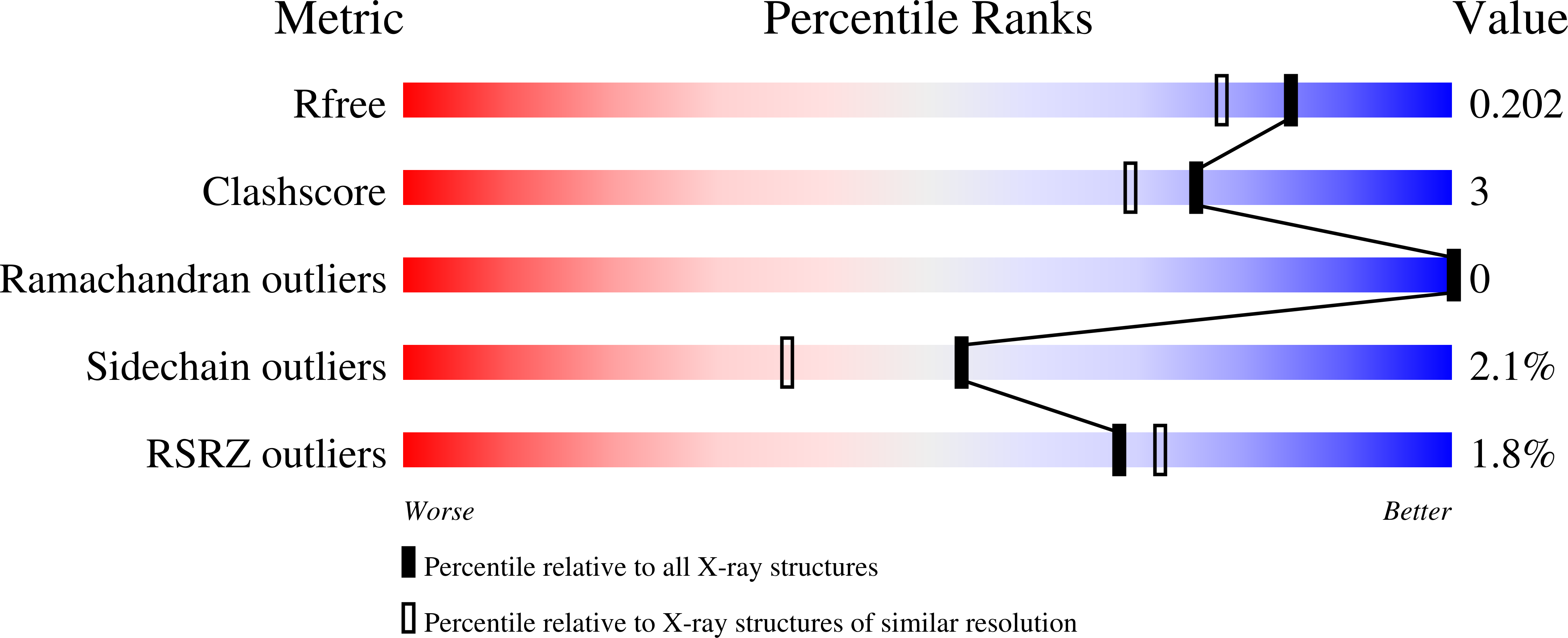
3SME - PubMed Abstract:
Hydrogen peroxide is a cell signaling agent that inactivates protein tyrosine phosphatases (PTPs) via oxidation of their catalytic cysteine residue. PTPs are inactivated rapidly during H(2)O(2)-mediated cellular signal transduction processes, but, paradoxically, hydrogen peroxide is a rather sluggish PTP inactivator in vitro. Here we present evidence that the biological buffer bicarbonate/CO(2) potentiates the ability of H(2)O(2) to inactivate PTPs. The results of biochemical experiments and high-resolution crystallographic analysis are consistent with a mechanism involving oxidation of the catalytic cysteine residue by peroxymonocarbonate generated via the reaction of H(2)O(2) with HCO(3)(-)/CO(2).
Organizational Affiliation:
Department of Chemistry, University of Missouri, Columbia, Missouri 65211, United States.