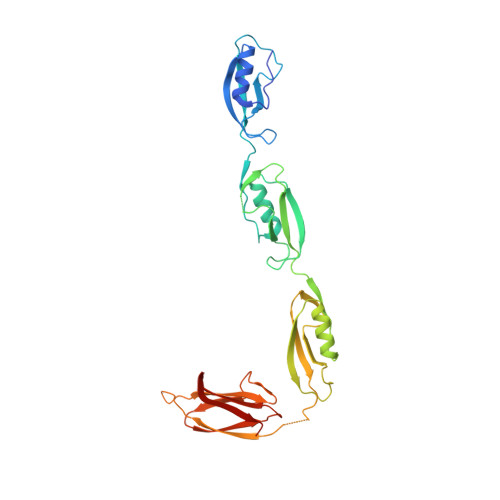
X-ray structural studies of the entire extracellular region of the serine/threonine kinase PrkC from Staphylococcus aureus.
Ruggiero, A., Squeglia, F., Marasco, D., Marchetti, R., Molinaro, A., Berisio, R.(2011) Biochem J 435: 33-41
- PubMed: 21208192
- DOI: https://doi.org/10.1042/BJ20101643
- Primary Citation of Related Structures:
3PY9 - PubMed Abstract:
Bacterial serine/threonine kinases modulate a wide number of cellular processes. The serine/threonine kinase PrkC from the human pathogen Staphylococcus aureus was also shown to induce germination of Bacillus subtilis spores, in response to cell wall muropeptides. The presence of muropeptides in the bacterial extracellular milieu is a strong signal that the growing conditions are promising. In the present paper, we report the X-ray structure of the entire extracellular region of PrkC from S. aureus. This structure reveals that the extracellular region of PrkC, EC-PrkC, is a linear modular structure composed of three PASTA (penicillin binding-associated and serine/threonine kinase-associated) domains and an unpredicted C-terminal domain, which presents the typical features of adhesive proteins. Using several solution techniques, we also found that EC-PrkC shows no tendency to dimerize even in the presence of high concentrations of muropeptides. X-ray structural results obtained in the present study provide molecular clues into the mechanism of muropeptide-induced PrkC activation.
Organizational Affiliation:
Institute of Biostructures and Bioimaging, CNR, Via Mezzocannone 16, I-80134, Napoli, Italy.