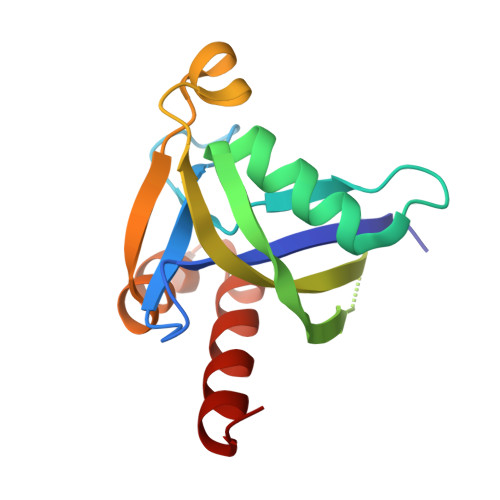
Crystal structure of human diphosphoinositol polyphosphate phosphohydrolase 3-alpha
Tresaugues, L., Welin, M., Arrowsmith, C.H., Berglund, H., Bountra, C., Collins, R., Edwards, A.M., Flodin, S., Flores, A., Graslund, S., Hammarstrom, M., Johansson, I., Karlberg, T., Kol, S., Kotenyova, T., Moche, M., Nyman, T., Persson, C., Schuler, H., Schutz, P., Siponen, M.I., Thorsell, A.G., van der Berg, S., Wahlberg, E., Weigelt, J., Wisniewska, M., Nordlund, P.To be published.