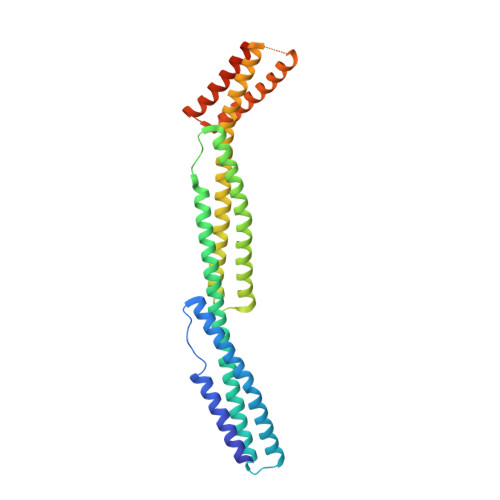
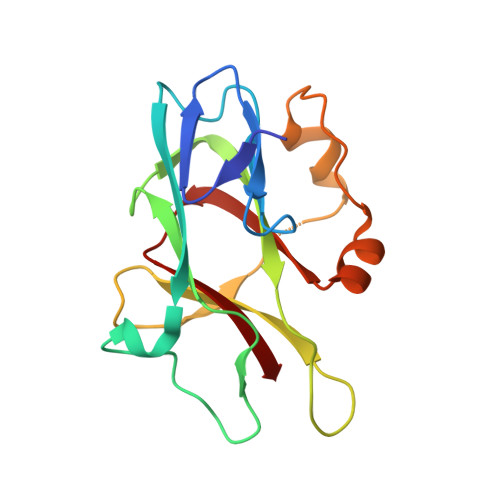
Structural basis for spectrin recognition by ankyrin.
Ipsaro, J.J., Mondragon, A.(2010) Blood 115: 4093-4101
- PubMed: 20101027
- DOI: https://doi.org/10.1182/blood-2009-11-255604
- Primary Citation of Related Structures:
3KBT, 3KBU - PubMed Abstract:
Maintenance of membrane integrity and organization in the metazoan cell is accomplished through intracellular tethering of membrane proteins to an extensive, flexible protein network. Spectrin, the principal component of this network, is anchored to membrane proteins through the adaptor protein ankyrin. To elucidate the atomic basis for this interaction, we determined a crystal structure of human betaI-spectrin repeats 13 to 15 in complex with the ZU5-ANK domain of human ankyrin R. The structure reveals the role of repeats 14 to 15 in binding, the electrostatic and hydrophobic contributions along the interface, and the necessity for a particular orientation of the spectrin repeats. Using structural and biochemical data as a guide, we characterized the individual proteins and their interactions by binding and thermal stability analyses. In addition to validating the structural model, these data provide insight into the nature of some mutations associated with cell morphology defects, including those found in human diseases such as hereditary spherocytosis and elliptocytosis. Finally, analysis of the ZU5 domain suggests it is a versatile protein-protein interaction module with distinct interaction surfaces. The structure represents not only the first of a spectrin fragment in complex with its binding partner, but also that of an intermolecular complex involving a ZU5 domain.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology, and Cell Biology, Northwestern University, Evanston, IL 60208, USA.