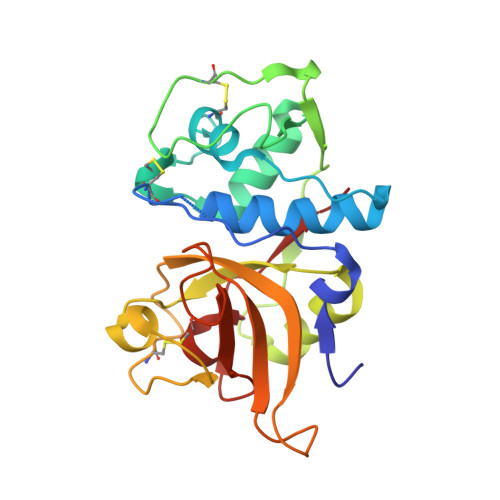
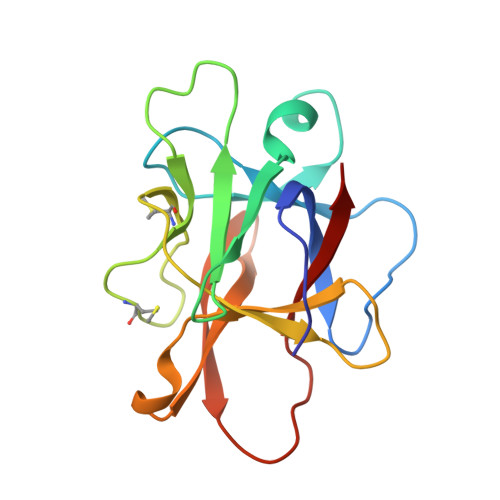
Versatile loops in mycocypins inhibit three protease families.
Renko, M., Sabotic, J., Mihelic, M., Brzin, J., Kos, J., Turk, D.(2010) J Biol Chem 285: 308-316
- PubMed: 19846555
- DOI: https://doi.org/10.1074/jbc.M109.043331
- Primary Citation of Related Structures:
3H6Q, 3H6R, 3H6S - PubMed Abstract:
Mycocypins, clitocypins and macrocypins, are cysteine protease inhibitors isolated from the mushrooms Clitocybe nebularis and Macrolepiota procera. Lack of sequence homology to other families of protease inhibitors suggested that mycocypins inhibit their target cysteine protease by a unique mechanism and that a novel fold may be found. The crystal structures of the complex of clitocypin with the papain-like cysteine protease cathepsin V and of macrocypin and clitocypin alone have revealed yet another motif of binding to papain like-cysteine proteases, which in a yet unrevealed way occludes the catalytic residue. The binding is associated with a peptide-bond flip of glycine that occurs before or concurrently with the inhibitor docking. Mycocypins possess a beta-trefoil fold, the hallmark of Kunitz-type inhibitors. It is a tree-like structure with two loops in the root region, a stem comprising a six-stranded beta-barrel, and two layers of loops (6 + 3) in the crown region. The two loops that bind to cysteine cathepsins belong to the lower layer of the crown loops, whereas a single loop from the crown region can inhibit trypsin or asparaginyl endopeptidase, as demonstrated by site-directed mutagenesis. These loops present a versatile surface with the potential to bind to additional classes of proteases. When appropriately engineered, they could provide the basis for possible exploitation in crop protection.
Organizational Affiliation:
Department of Biochemistry and Molecular, Jozef Stefan Institute,Jamova 39, 1000 Ljubljana, Slovenia.