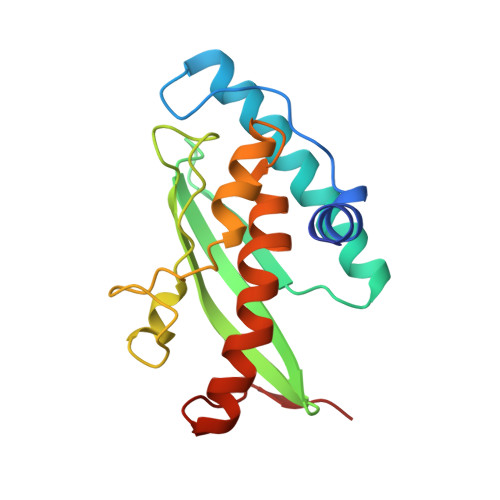
Crystal structure of Ufc1, the Ufm1-conjugating enzyme
Mizushima, T., Tatsumi, K., Ozaki, Y., Kawakami, T., Suzuki, A., Ogasahara, K., Komatsu, M., Kominami, E., Tanaka, K., Yamane, T.(2007) Biochem Biophys Res Commun 362: 1079-1084
- PubMed: 17825256
- DOI: https://doi.org/10.1016/j.bbrc.2007.08.129
- Primary Citation of Related Structures:
2Z6O, 2Z6P - PubMed Abstract:
Ubiquitin and ubiquitin-like protein-conjugating enzymes play central roles in posttranslational modification processes. The ubiquitin-fold modifier 1 (Ufm1), one of a variety of ubiquitin-like modifiers, is covalently attached to target proteins via Uba5 and Ufm1-conjugating enzyme 1 (Ufc1), which are analogous to the E1 and E2 ubiquitylation enzymes. As Ufm1-related proteins are conserved in metazoa and plants, the Ufm1 system likely plays important roles in various multicellular organisms. Herein, we report the X-ray structure of human Ufc1 determined at 1.6 A resolution. The Ufc1 structure comprises a canonical E2 domain and an additional N-terminal domain. The Uba5 binding site on Ufc1 was assigned by structural comparison of Ufc1 and Ubc12 and related mutational analyses. In addition, we show that the N-terminal unique domain of Ufc1 contributes to thermal stability.
Organizational Affiliation:
Department of Biotechnology, Graduate School of Engineering, Nagoya University, Chikusa-ku, Nagoya 464-8603, Japan.