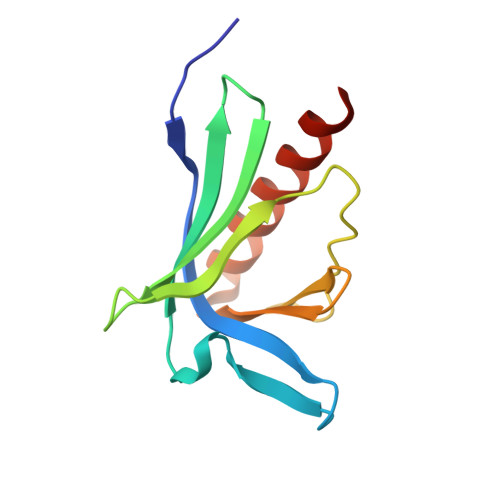
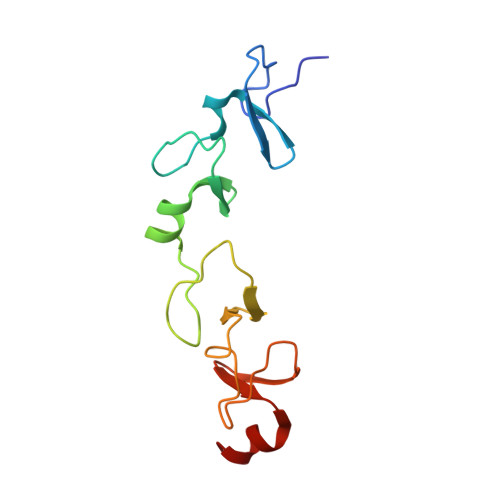
Molecular recognition of the Tes LIM2-3 domains by the actin-related protein Arp7A.
Boeda, B., Knowles, P.P., Briggs, D.C., Murray-Rust, J., Soriano, E., Garvalov, B.K., McDonald, N.Q., Way, M.(2011) J Biol Chem 286: 11543-11554
- PubMed: 21278383
- DOI: https://doi.org/10.1074/jbc.M110.171264
- Primary Citation of Related Structures:
2XQN - PubMed Abstract:
Actin-related proteins (Arps) are a highly conserved family of proteins that have extensive sequence and structural similarity to actin. All characterized Arps are components of large multimeric complexes associated with chromatin or the cytoskeleton. In addition, the human genome encodes five conserved but largely uncharacterized "orphan" Arps, which appear to be mostly testis-specific. Here we show that Arp7A, which has 43% sequence identity with β-actin, forms a complex with the cytoskeletal proteins Tes and Mena in the subacrosomal layer of round spermatids. The N-terminal 65-residue extension to the actin-like fold of Arp7A interacts directly with Tes. The crystal structure of the 1-65(Arp7A)·LIM2-3(Tes)·EVH1(Mena) complex reveals that residues 28-49 of Arp7A contact the LIM2-3 domains of Tes. Two alanine residues from Arp7A that occupy equivalent apolar pockets in both LIM domains as well as an intervening GPAK linker that binds the LIM2-3 junction are critical for the Arp7A-Tes interaction. Equivalent occupied apolar pockets are also seen in the tandem LIM domain structures of LMO4 and Lhx3 bound to unrelated ligands. Our results indicate that apolar pocket interactions are a common feature of tandem LIM domain interactions, but ligand specificity is principally determined by the linker sequence.
Organizational Affiliation:
Cell Motility Laboratory, Cancer Research UK, London Research Institute, London, United Kingdom.