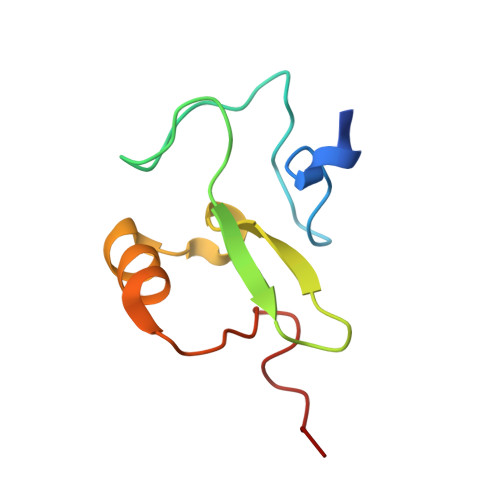
Rag2 Phd Finger Couples Histone H3 Lysine 4 Trimethylation with V(D)J Recombination.
Matthews, A.G.W., Kuo, A.J., Ramon-Maiques, S., Han, S., Champagne, K.S., Ivanov, D., Gallardo, M., Carney, D., Cheung, P., Ciccone, D.N., Walter, K.L., Utz, P.J., Shi, Y., Kutateladze, T.G., Yang, W., Gozani, O., Oettinger, M.A.(2007) Nature 450: 1106
- PubMed: 18033247
- DOI: https://doi.org/10.1038/nature06431
- Primary Citation of Related Structures:
2V89 - PubMed Abstract:
Nuclear processes such as transcription, DNA replication and recombination are dynamically regulated by chromatin structure. Eukaryotic transcription is known to be regulated by chromatin-associated proteins containing conserved protein domains that specifically recognize distinct covalent post-translational modifications on histones. However, it has been unclear whether similar mechanisms are involved in mammalian DNA recombination. Here we show that RAG2--an essential component of the RAG1/2 V(D)J recombinase, which mediates antigen-receptor gene assembly--contains a plant homeodomain (PHD) finger that specifically recognizes histone H3 trimethylated at lysine 4 (H3K4me3). The high-resolution crystal structure of the mouse RAG2 PHD finger bound to H3K4me3 reveals the molecular basis of H3K4me3-recognition by RAG2. Mutations that abrogate RAG2's recognition of H3K4me3 severely impair V(D)J recombination in vivo. Reducing the level of H3K4me3 similarly leads to a decrease in V(D)J recombination in vivo. Notably, a conserved tryptophan residue (W453) that constitutes a key structural component of the K4me3-binding surface and is essential for RAG2's recognition of H3K4me3 is mutated in patients with immunodeficiency syndromes. Together, our results identify a new function for histone methylation in mammalian DNA recombination. Furthermore, our results provide the first evidence indicating that disrupting the read-out of histone modifications can cause an inherited human disease.
Organizational Affiliation:
Department of Molecular Biology, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts 02114, USA.