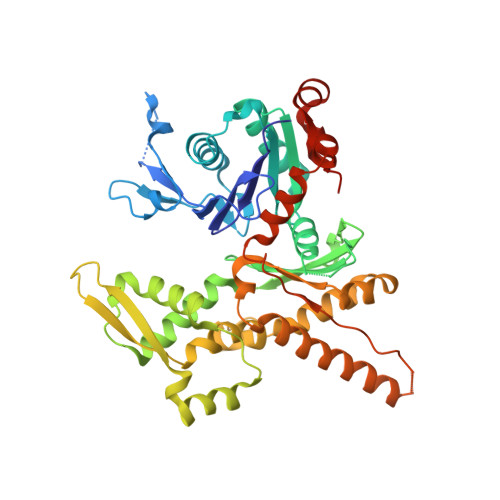
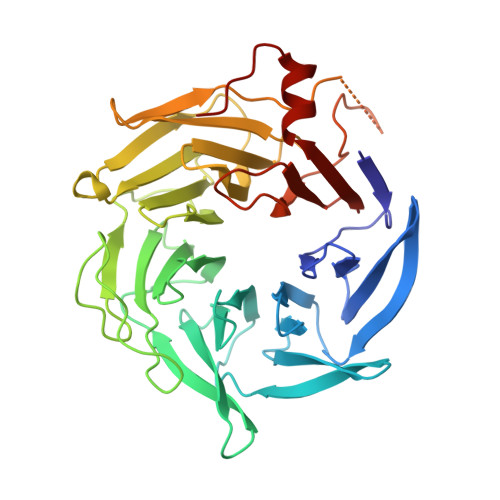
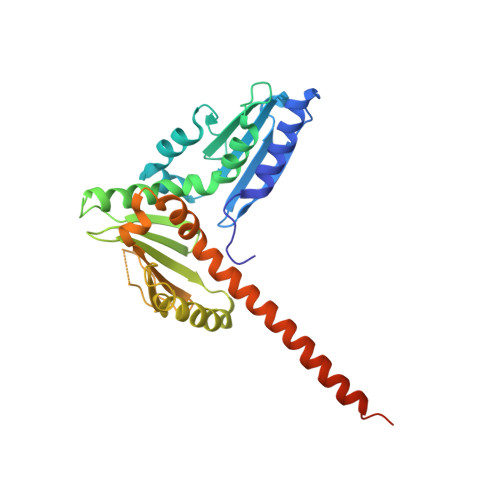
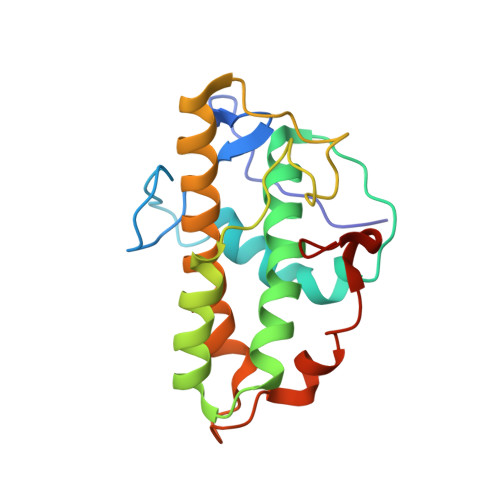
Insights into the Influence of Nucleotides on Actin Family Proteins from Seven Structures of Arp2/3 Complex.
Nolen, B.J., Pollard, T.D.(2007) Mol Cell 26: 449-457
- PubMed: 17499050
- DOI: https://doi.org/10.1016/j.molcel.2007.04.017
- Primary Citation of Related Structures:
2P9I, 2P9K, 2P9L, 2P9N, 2P9P, 2P9S, 2P9U - PubMed Abstract:
ATP is required for nucleation of actin filament branches by Arp2/3 complex, but the influence of ATP binding and hydrolysis are poorly understood. We determined crystal structures of bovine Arp2/3 complex cocrystallized with various bound adenine nucleotides and cations. Nucleotide binding favors closure of the nucleotide-binding cleft of Arp3, but no large-scale conformational changes in the complex. Thus, ATP binding does not directly activate Arp2/3 complex but is part of a network of interactions that contribute to nucleation. We compared nucleotide-induced conformational changes of residues lining the cleft in Arp3 and actin structures to construct a movie depicting the proposed ATPase cycle for the actin family. Chemical crosslinking stabilized subdomain 1 of Arp2, revealing new electron density for 69 residues in this subdomain. Steric clashes with Arp3 appear to be responsible for intrinsic disorder of subdomains 1 and 2 of Arp2 in inactive Arp2/3 complex.
Organizational Affiliation:
Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06520-8103, USA.