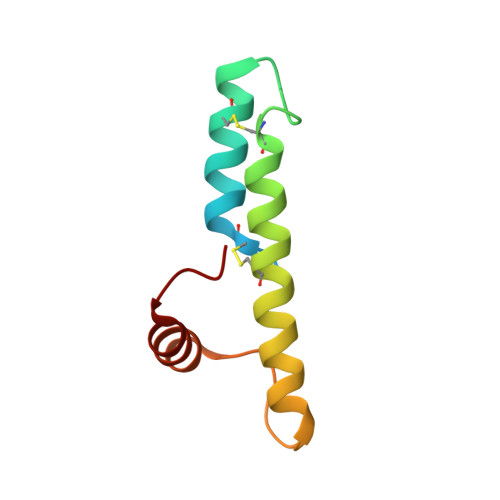
Structural characterization of CHCHD5 and CHCHD7: Two atypical human twin CX(9)C proteins.
Banci, L., Bertini, I., Ciofi-Baffoni, S., Jaiswal, D., Neri, S., Peruzzini, R., Winkelmann, J.(2012) J Struct Biol 180: 190-200
- PubMed: 22842048
- DOI: https://doi.org/10.1016/j.jsb.2012.07.007
- Primary Citation of Related Structures:
2LQL, 2LQT - PubMed Abstract:
Twin CX(9)C proteins constitute a large protein family among all eukaryotes; are putative substrates of the mitochondrial Mia40-dependent import machinery; contain a coiled coil-helix-coiled coil-helix (CHCH) fold stabilized by two disulfide bonds as exemplified by three structures available for this family. However, they considerably differ at the primary sequence level and this prevents an accurate prediction of their structural models. With the aim of expanding structural information on CHCH proteins, here we structurally characterized human CHCHD5 and CHCHD7. While CHCHD5 has two weakly interacting CHCH domains which sample a range of limited conformations as a consequence of hydrophobic interactions, CHCHD7 has a third helix hydrophobically interacting with an extension of helix α2, which is part of the CHCH domain. Upon reduction of the disulfide bonds both proteins become unstructured exposing hydrophobic patches, with the result of protein aggregation/precipitation. These results suggest a model where the molecular interactions guiding the protein recognition between Mia40 and the disulfide-reduced CHCHD5 and CHCHD7 substrates occurs in vivo when the latter proteins are partially embedded in the protein import pore of the outer membrane of mitochondria.
Organizational Affiliation:
Magnetic Resonance Center CERM, University of Florence, Via Luigi Sacconi 6, 50019 Sesto Fiorentino, Florence, Italy. banci@cerm.unifi.it