Solution structure of human zeta-COP: direct evidences for structural similarity between COP I and clathrin-adaptor coats
Yu, W., Lin, J., Jin, C., Xia, B.(2009) J Mol Biol 386: 903-912
- PubMed: 19167404
- DOI: https://doi.org/10.1016/j.jmb.2008.12.083
- Primary Citation of Related Structures:
2HF6 - PubMed Abstract:
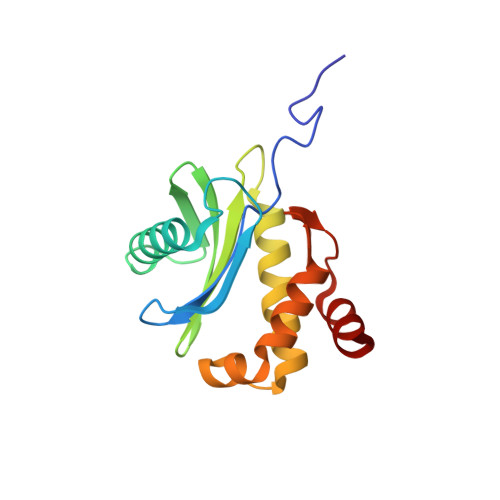
COP-I-coated vesicles are protein and lipid carriers that mediate intra-Golgi transport and transport from the cis-Golgi complex to the endoplasmic reticulum in cells. The coatomer of the vesicles coat is comprised of seven subunits: alpha-COP, epsilon-COP, beta'-COP, beta-COP, gamma-COP, delta-COP, and zeta-COP. Here we report the solution structure of a truncated form (residues 1-149; zeta-COP149) of human zeta-COP (total 177 residues). It is the first three-dimensional structure of a "core" subunit of the COP I F-subcomplex. The structure of zeta-COP149 mainly consists of a disordered N-terminal tail, a five-stranded antiparallel beta-sheet, a two-stranded antiparallel beta-sheet, and five alpha-helices. The global folding of zeta-COP149 is very similar to the crystal structures of AP1-sigma1 and AP2-sigma2, directly demonstrating the structural similarity between the "core" subunits of the COP I F-subcomplex and adaptor protein complexes. Through structural comparison and mutagenesis study, we have also demonstrated that the heterodimers of zeta-COP149 and gamma-COP have packing interfaces and relative subunit orientations similar to those of AP2-sigma2 and AP2-alpha heterodimers. These results provide direct evidence supporting the previous proposal that the COP I F-subcomplex and adaptor protein complexes have similar tertiary and quaternary structures.
Organizational Affiliation:
Beijing Nuclear Magnetic Resonance Center, Peking University, Beijing 100871, China.