Link between a novel human gamma-D-crystallin allele and a unique cataract phenotype explained by protein crystallography.
Kmoch, S., Brynda, J., Awsav, B., Bezouska, K., Novak, P., Rezacova, P., Ondrova, L., Filipec, M., Sedlacek, J., Elleder, M.(2000) Hum Mol Genet 12: 1779-1786
- PubMed: 10915766
- DOI: https://doi.org/10.1093/hmg/9.12.1779
- Primary Citation of Related Structures:
2G98 - PubMed Abstract:
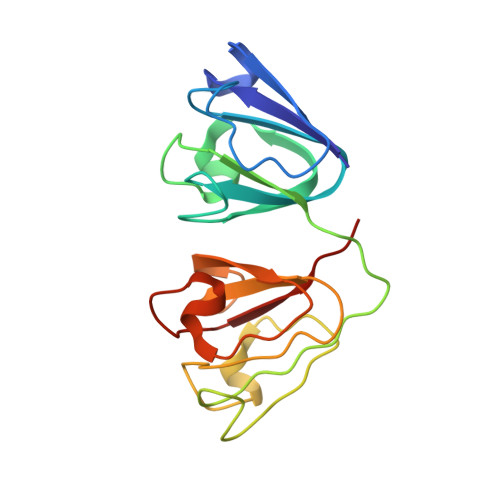
We describe a 5-year-old boy with a unique congenital cataract caused by deposition of numerous birefringent, pleiochroic and macroscopically prismatic crystals. Crystal analysis with subsequent automatic Edman degradation and matrix-associated laser desorption ionization time-of-flight mass spectrometry have identified the crystal-forming protein as gammaD-crystallin (CRYGD) lacking the N-terminal methionine. Sequencing of the CRYGD gene has shown a heterozygous C-->A transversion in position 109 of the inferred cDNA (36R-->S transversion of the processed, N-terminal methionine-lacking CRYGD). The lens protein crystals were X-ray diffracting, and our crystal structure solution at 2.25 A suggests that mutant R36S CRYGD has an unaltered protein fold. In contrast, the observed crystal packing is possible only with the mutant protein molecules that lack the bulky Arg36 side chain. This is the first described case of human cataract caused by crystallization of a protein in the lens. It involves the third known mutation in the CRYGD gene but offers, for the first time, a causative explanation of the phenotype.
Organizational Affiliation:
Institute of Inherited Metabolic Diseases, Division B, Building D, Ke Karlovu 2, 12808 Prague 2, Czech Republic.