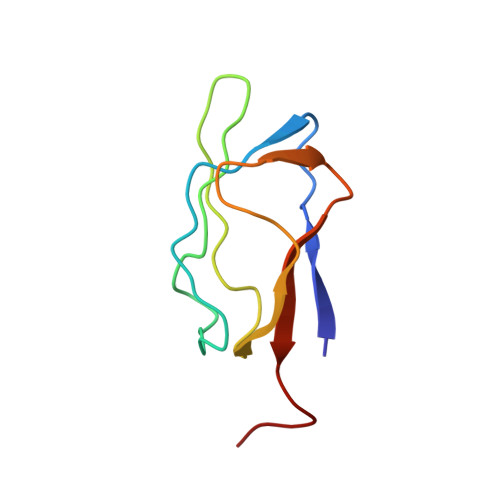
Three-dimensional structure in solution of the N-terminal lipoyl domain of the pyruvate dehydrogenase complex from Azotobacter vinelandii.
Berg, A., Vervoort, J., de Kok, A.(1997) Eur J Biochem 244: 352-360
- PubMed: 9119000
- DOI: https://doi.org/10.1111/j.1432-1033.1997.00352.x
- Primary Citation of Related Structures:
1IYU, 1IYV - PubMed Abstract:
The three-dimensional structure of the N-terminal lipoyl domain of the acetyltransferase component of the pyruvate dehydrogenase complex from Azotobacter vinelandii has been determined using heteronuclear multidimensional NMR spectroscopy and dynamical simulated annealing. The structure is compared with the solution structure of the lipoyl domain of the A. vinelandii 2-oxoglutarate dehydrogenase complex. The overall fold of the two structures, described as a beta-barrel-sandwich hybrid, is very similar. This agrees well with the high similarity of NMR-derived parameters, e.g. chemical shifts, between the two lipoyl domains. The main structural differences between the two lipoyl domains occur in a solvent-exposed loop close in space to the lipoylation site. Despite their high structural similarity, these lipoyl domains show a high preference for being reductively acylated by their parent 2-oxo acid dehydrogenase. Potential residues of the lipoyl domain involved in this process of molecular recognition are discussed.
Organizational Affiliation:
Department of Biochemistry, Agricultural University, Wageningen, The Netherlands.