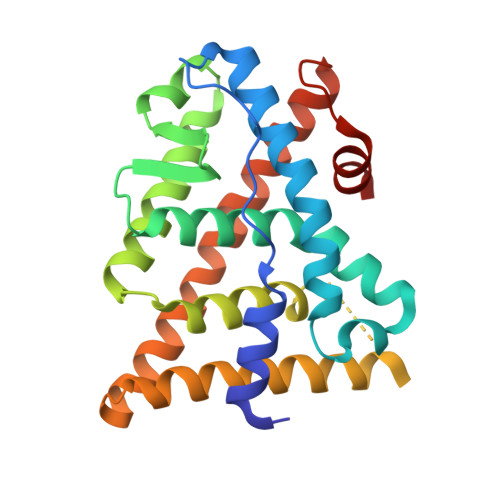
Understanding the Selectivity of Genistein for Human Estrogen Receptor-Beta Using X-Ray Crystallography and Computational Methods
Manas, E.S., Xu, Z.B., Unwalla, R.J., Somers, W.S.(2004) Structure 12: 2197-2207
- PubMed: 15576033
- DOI: https://doi.org/10.1016/j.str.2004.09.015
- Primary Citation of Related Structures:
1X7J, 1X7R - PubMed Abstract:
We present X-ray crystallographic and molecular modeling studies of estrogen receptors-alpha and -beta complexed with the estrogen receptor-beta-selective phytoestrogen genistein, and coactivator-derived NR box peptides containing an LXXLL motif. We demonstrate that the ligand binding mode is essentially identical when genistein is bound to both isoforms, despite the considerably weaker affinity of this ligand for estrogen receptor-alpha. In addition, we examine subtle differences between binding site residues, providing an explanation for why genistein is modestly selective for the beta isoform. To this end, we also present the results of quantum chemical studies and thermodynamic arguments that yield insight to the nature of the interactions leading to estrogen receptor-beta selectivity. The importance of our analysis to structure-based drug design is discussed.
Organizational Affiliation:
Department of Chemical and Screening Sciences, Wyeth Research, 500 Arcola Road, Collegeville, PA 19426, USA. manase@wyeth.com