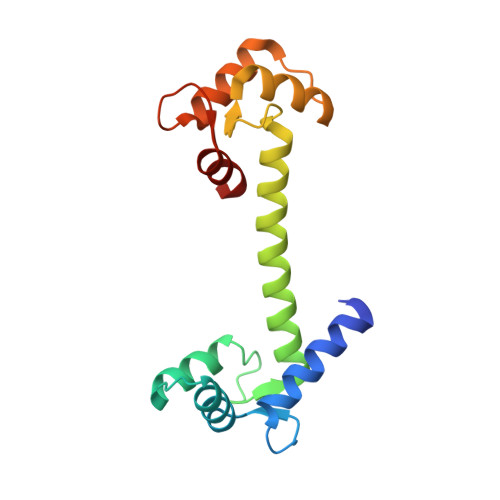
Crystal structure of human calmodulin-like protein: insights into its functional role.
Han, B.G., Han, M., Sui, H., Yaswen, P., Walian, P.J., Jap, B.K.(2002) FEBS Lett 521: 24-30
- PubMed: 12067719
- DOI: https://doi.org/10.1016/s0014-5793(02)02780-1
- Primary Citation of Related Structures:
1GGZ - PubMed Abstract:
A calmodulin (CaM)-like protein (hCLP) is expressed in human mammary epithelial cells but appears to be limited to certain epithelial cells such as those found in skin, prostate, breast and cervical tissues. A decrease in the expression of this protein is associated with the occurrence of tumors in breast epithelium. The structure of hCLP determined to 1.5 A resolution by X-ray crystallography shows a distinct 30 degrees displacement along the interconnecting central helix, when compared to the highly conserved structure of vertebrate CaM, resulting in a difference in the relative orientation of its two globular domains. Additionally, the electric surface potential landscape at the target protein binding regions on the two globular domains of hCLP is significantly different from those of CaM, indicating that the respective ranges of hCLP and hCaM target proteins do not fully overlap. Observations that hCLP can competitively inhibit CaM activation of target proteins also imply a role for hCLP in which it may also serve as a modulator of CaM activity in the epithelial cells where hCLP is expressed.
Organizational Affiliation:
Life Sciences Division, Lawrence Berkeley National Laboratory, University of California, Berkeley, CA 94720, USA.