Structural Insights Into the High Selectivity of the Anti-Diabetic Drug Mitiglinide
Wang, M.M., Wu, J.X., Chen, L.(2022) Front Pharmacol 13: 929684-929684
- PubMed: 35847046
- DOI: https://doi.org/10.3389/fphar.2022.929684
- Primary Citation of Related Structures:
7WIT - PubMed Abstract:
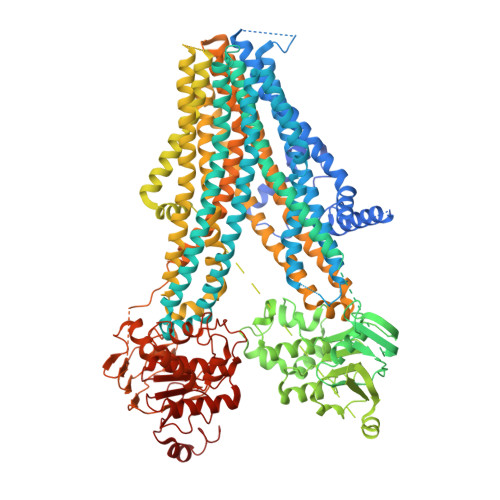
Mitiglinide is a highly selective fast-acting anti-diabetic drug that induces insulin secretion by inhibiting pancreatic K ATP channels. However, how mitiglinide binds K ATP channels remains unknown. Here, we show the cryo-EM structure of the SUR1 subunit complexed with mitiglinide. The structure reveals that mitiglinide binds inside the common insulin secretagogue-binding site of SUR1, which is surrounded by TM7, TM8, TM16, and TM17. Mitiglinide locks SUR1 in the NBD-separated inward-facing conformation. The detailed structural analysis of the mitiglinide-binding site uncovers the molecular basis of its high selectivity.
Organizational Affiliation:
State Key Laboratory of Membrane Biology, Beijing Key Laboratory of Cardiometabolic Molecular Medicine, College of Future Technology, Institute of Molecular Medicine, Peking University, Beijing, China.