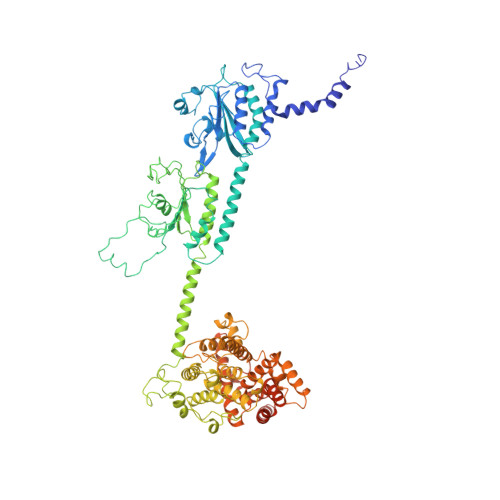
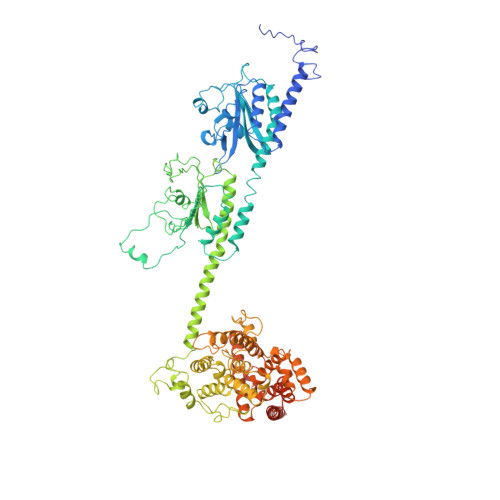
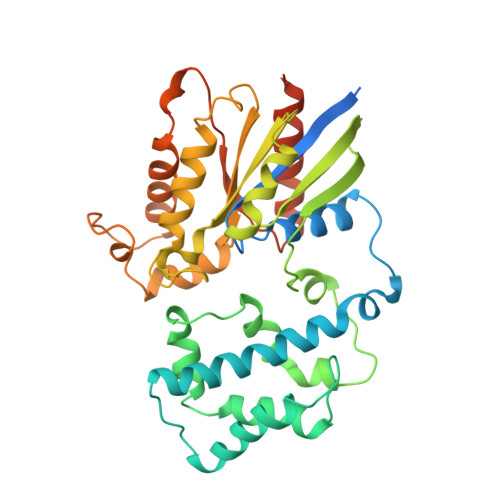
Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6.
Gao, Y., Eskici, G., Ramachandran, S., Poitevin, F., Seven, A.B., Panova, O., Skiniotis, G., Cerione, R.A.(2020) Mol Cell 80: 237
- PubMed: 33007200
- DOI: https://doi.org/10.1016/j.molcel.2020.09.013
- Primary Citation of Related Structures:
7JSN - PubMed Abstract:
Heterotrimeric G proteins communicate signals from activated G protein-coupled receptors to downstream effector proteins. In the phototransduction pathway responsible for vertebrate vision, the G protein-effector complex is composed of the GTP-bound transducin α subunit (Gα T ·GTP) and the cyclic GMP (cGMP) phosphodiesterase 6 (PDE6), which stimulates cGMP hydrolysis, leading to hyperpolarization of the photoreceptor cell. Here we report a cryo-electron microscopy (cryoEM) structure of PDE6 complexed to GTP-bound Gα T . The structure reveals two Gα T ·GTP subunits engaging the PDE6 hetero-tetramer at both the PDE6 catalytic core and the PDEγ subunits, driving extensive rearrangements to relieve all inhibitory constraints on enzyme catalysis. Analysis of the conformational ensemble in the cryoEM data highlights the dynamic nature of the contacts between the two Gα T ·GTP subunits and PDE6 that supports an alternating-site catalytic mechanism.
Organizational Affiliation:
Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA 94305, USA; Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94305, USA.