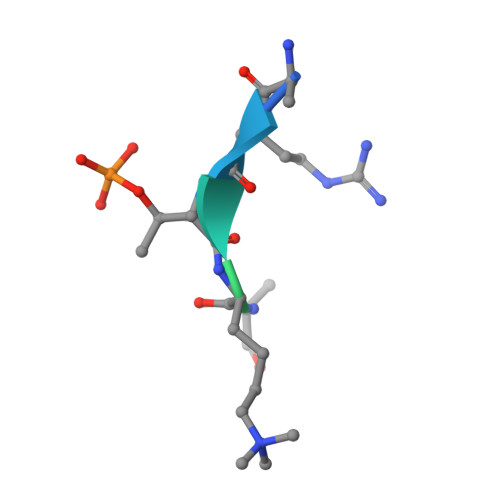
Tip60 acetylation of histone H3K4 temporally controls chromosome passenger complex localization.
Niedzialkowska, E., Liu, L., Kuscu, C., Mayo, Z., Minor, W., Strahl, B.D., Adli, M., Stukenberg, P.T.(2022) Mol Biol Cell 33: br15-br15
- PubMed: 35653296
- DOI: https://doi.org/10.1091/mbc.E21-06-0283
- Primary Citation of Related Structures:
7LBK, 7LBO, 7LBP, 7LBQ - PubMed Abstract:
The Chromosome Passenger Complex (CPC) generates chromosome autonomous signals that regulate mitotic events critical for genome stability. Tip60 is a lysine acetyltransferase that is a tumor suppressor and is targeted for proteasomal degradation by oncogenic papilloma viruses. Mitotic regulation requires the localization of the CPC to inner centromeres, which is driven by the Haspin kinase phosphorylating histone H3 on threonine 3 (H3T3ph). Here we describe how Tip60 acetylates histone H3 at lysine 4 (H3K4ac) to block both the H3T3ph writer and the reader to ensure that this mitotic signaling cannot begin before prophase. Specifically, H3K4ac inhibits Haspin phosphorylation of H3T3 and prevents binding of the Survivin subunit to H3T3ph. Tip60 acetylates H3K4 during S/G2 at centromeres. Inhibition of Tip60 allows the CPC to bind centromeres in G2 cells, and targeting of Tip60 to centromeres prevents CPC localization in mitosis. The H3K4ac mark is removed in prophase by HDAC3 to initiate the CPC localization cascade. Together, our results suggest that Tip60 and HDAC3 temporally control H3K4 acetylation to precisely time the targeting of the CPC to inner centromeres.
Organizational Affiliation:
Department of Biochemistry and Molecular Genetics, University of Virginia, Charlottesville VA 22908.