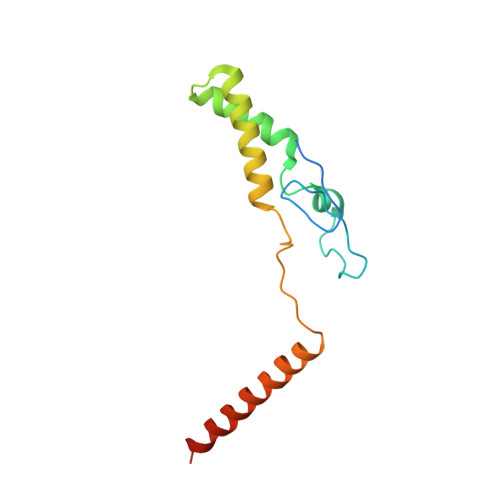
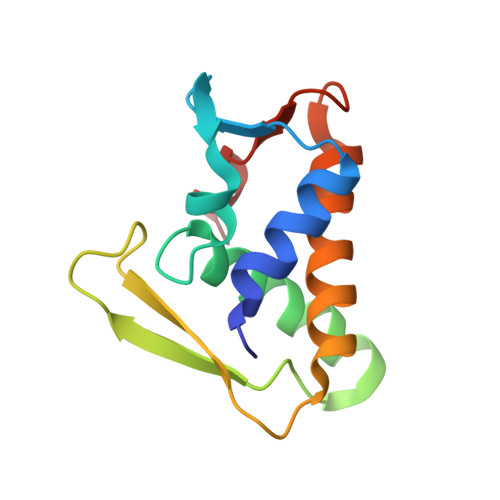
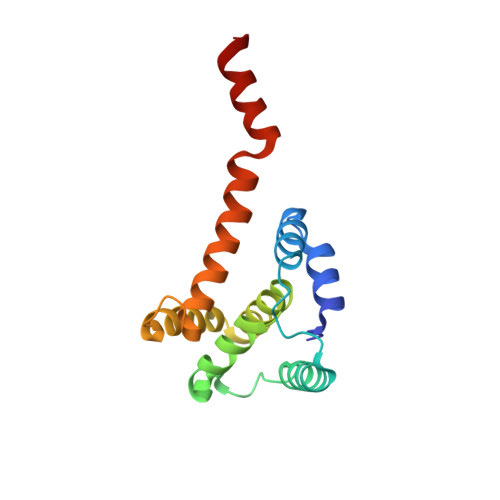
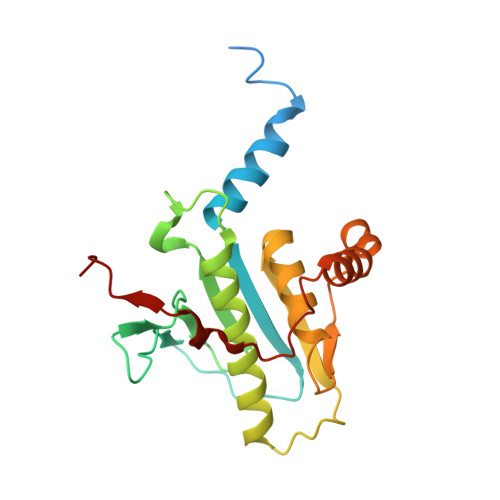
Structural basis for effector protein recognition by the Dot/Icm Type IVB coupling protein complex.
Kim, H., Kubori, T., Yamazaki, K., Kwak, M.J., Park, S.Y., Nagai, H., Vogel, J.P., Oh, B.H.(2020) Nat Commun 11: 2623-2623
- PubMed: 32457311
- DOI: https://doi.org/10.1038/s41467-020-16397-0
- Primary Citation of Related Structures:
7BWK - PubMed Abstract:
The Legionella pneumophila Dot/Icm type IVB secretion system (T4BSS) is extremely versatile, translocating ~300 effector proteins into host cells. This specialized secretion system employs the Dot/Icm type IVB coupling protein (T4CP) complex, which includes IcmS, IcmW and LvgA, that are known to selectively assist the export of a subclass of effectors. Herein, the crystal structure of a four-subunit T4CP subcomplex bound to the effector protein VpdB reveals an interaction between LvgA and a linear motif in the C-terminus of VpdB. The same binding interface of LvgA also interacts with the C-terminal region of three additional effectors, SidH, SetA and PieA. Mutational analyses identified a FxxxLxxxK binding motif that is shared by VpdB and SidH, but not by SetA and PieA, showing that LvgA recognizes more than one type of binding motif. Together, this work provides a structural basis for how the Dot/Icm T4CP complex recognizes effectors, and highlights the multiple substrate-binding specificities of its adaptor subunit.
Organizational Affiliation:
Department of Biological Sciences, KAIST Institute for the Biocentury, Korea Advanced Institute of Science and Technology, Daejeon, 34141, Republic of Korea.