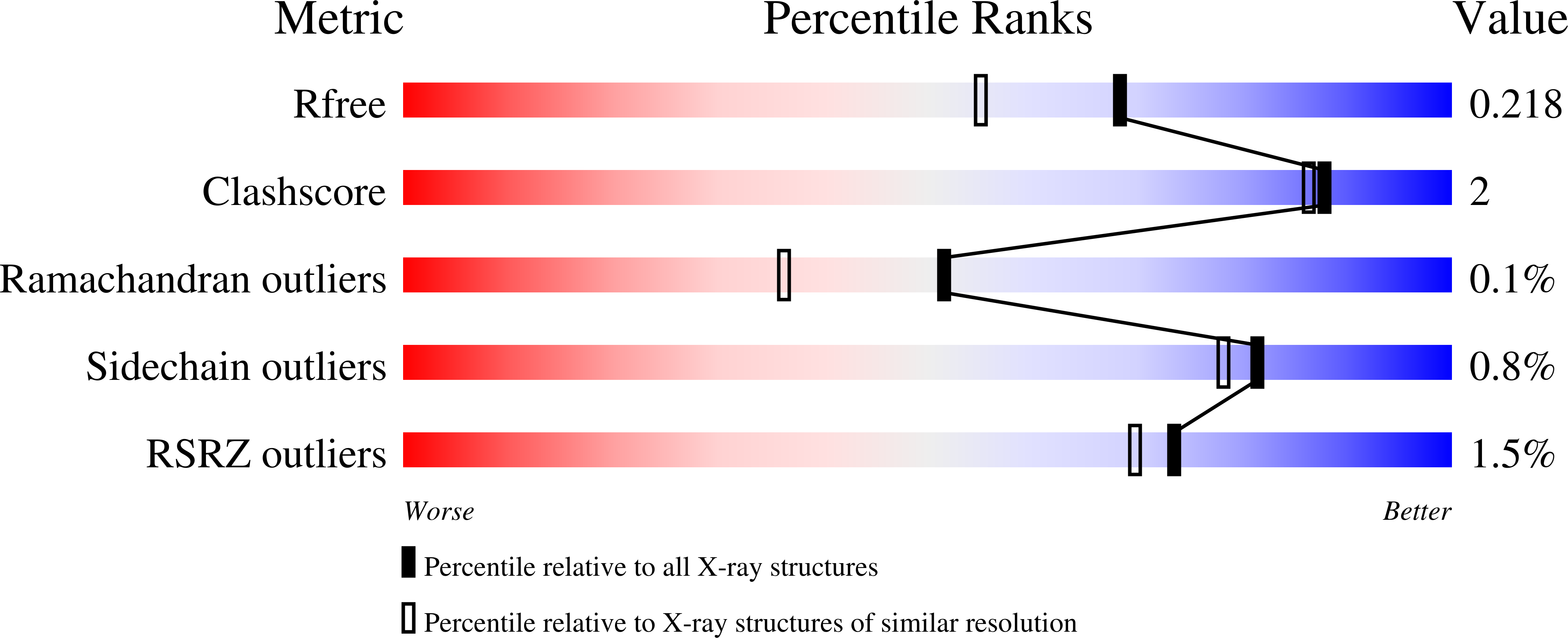
ACE-domain selectivity extends beyond direct interacting residues at the active site.
Cozier, G.E., Lubbe, L., Sturrock, E.D., Acharya, K.R.(2020) Biochem J 477: 1241-1259
- PubMed: 32195541
- DOI: https://doi.org/10.1042/BCJ20200060
- Primary Citation of Related Structures:
6TT1, 6TT3, 6TT4 - PubMed Abstract:
Angiotensin-converting enzyme (ACE) is best known for its formation of the vasopressor angiotensin II that controls blood pressure but is also involved in other physiological functions through the hydrolysis of a variety of peptide substrates. The enzyme contains two catalytic domains (nACE and cACE) that have different affinities for ACE substrates and inhibitors. We investigated whether nACE inhibitor backbones contain a unique property which allows them to take advantage of the hinging of nACE. Kinetic analysis showed that mutation of unique nACE residues, in both the S2 pocket and around the prime subsites (S') to their C-domain counterparts, each resulted in a decrease in the affinity of nACE specific inhibitors (SG6, 33RE and ketoACE-13) but it required the combined S2_S' mutant to abrogate nACE-selectivity. However, this was not observed with the non-domain-selective inhibitors enalaprilat and omapatrilat. High-resolution structures were determined for the minimally glycosylated nACE with the combined S2_S' mutations in complex with the ACE inhibitors 33RE (1.8 Å), omapatrilat (1.8 Å) and SG6 (1.7 Å). These confirmed that the affinities of the nACE-selective SG6, 33RE and ketoACE-13 are not only affected by direct interactions with the immediate environment of the binding site, but also by more distal residues. This study provides evidence for a more general mechanism of ACE inhibition involving synergistic effects of not only the S2, S1' and S2' subsites, but also residues involved in the sub-domain interface that effect the unique ways in which the two domains stabilize active site loops to favour inhibitor binding.
Organizational Affiliation:
Department of Biology and Biochemistry, University of Bath, Claverton Down, Bath BA2 7AY, U.K.