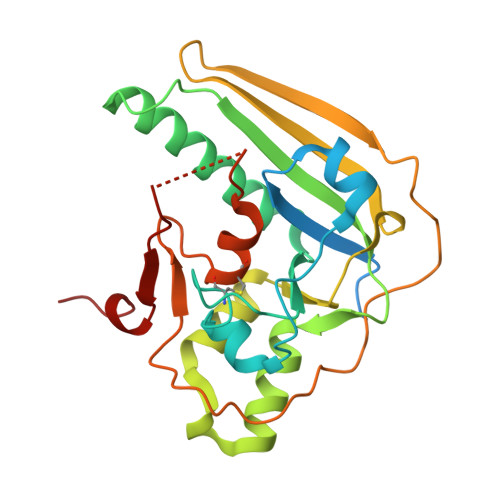
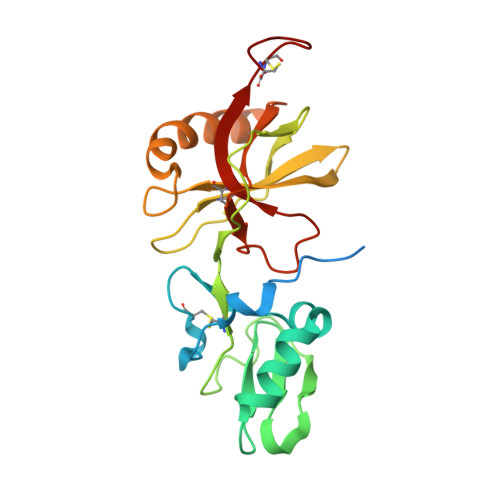
Genetically detoxified pertussis toxin displays near identical structure to its wild-type and exhibits robust immunogenicity.
Ausar, S.F., Zhu, S., Duprez, J., Cohen, M., Bertrand, T., Steier, V., Wilson, D.J., Li, S., Sheung, A., Brookes, R.H., Pedyczak, A., Rak, A., Andrew James, D.(2020) Commun Biol 3: 427-427
- PubMed: 32759959
- DOI: https://doi.org/10.1038/s42003-020-01153-3
- Primary Citation of Related Structures:
6RO0 - PubMed Abstract:
The mutant gdPT R9K/E129G is a genetically detoxified variant of the pertussis toxin (PTx) and represents an attractive candidate for the development of improved pertussis vaccines. The impact of the mutations on the overall protein structure and its immunogenicity has remained elusive. Here we present the crystal structure of gdPT and show that it is nearly identical to that of PTx. Hydrogen-deuterium exchange mass spectrometry revealed dynamic changes in the catalytic domain that directly impacted NAD + binding which was confirmed by biolayer interferometry. Distal changes in dynamics were also detected in S2-S5 subunit interactions resulting in tighter packing of B-oligomer corresponding to increased thermal stability. Finally, antigen stimulation of human whole blood, analyzed by a previously unreported mass cytometry assay, indicated broader immunogenicity of gdPT compared to pertussis toxoid. These findings establish a direct link between the conserved structure of gdPT and its ability to generate a robust immune response.
Organizational Affiliation:
Bioprocess Research and Development, Sanofi Pasteur Ltd., Toronto, ON, M2R 3T4, Canada.