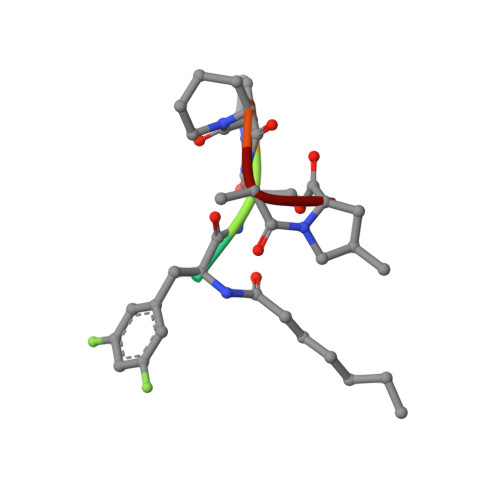
Ureadepsipeptides as ClpP Activators.
Griffith, E.C., Zhao, Y., Singh, A.P., Conlon, B.P., Tangallapally, R., Shadrick, W.R., Liu, J., Wallace, M.J., Yang, L., Elmore, J.M., Li, Y., Zheng, Z., Miller, D.J., Cheramie, M.N., Lee, R.B., LaFleur, M.D., Lewis, K., Lee, R.E.(2019) ACS Infect Dis 5: 1915-1925
- PubMed: 31588734
- DOI: https://doi.org/10.1021/acsinfecdis.9b00245
- Primary Citation of Related Structures:
5VZ2, 5W18, 6PKA, 6PMD - PubMed Abstract:
Acyldepsipeptides are a unique class of antibiotics that act via allosterically dysregulated activation of the bacterial caseinolytic protease (ClpP). The ability of ClpP activators to kill nongrowing bacteria represents a new opportunity to combat deep-seated biofilm infections. However, the acyldepsipeptide scaffold is subject to rapid metabolism. Herein, we explore alteration of the potentially metabolically reactive α,β unsaturated acyl chain. Through targeted synthesis, a new class of phenyl urea substituted depsipeptide ClpP activators with improved metabolic stability is described. The ureadepsipeptides are potent activators of Staphylococcus aureus ClpP and show activity against Gram-positive bacteria, including S. aureus biofilms. These studies demonstrate that a phenyl urea motif can successfully mimic the double bond, maintaining potency equivalent to acyldepsipeptides but with decreased metabolic liability. Although removal of the double bond from acyldepsipeptides generally has a significant negative impact on potency, structural studies revealed that the phenyl ureadepsipeptides can retain potency through the formation of a third hydrogen bond between the urea and the key Tyr63 residue in the ClpP activation domain. Ureadepsipeptides represent a new class of ClpP activators with improved drug-like properties, potent antibacterial activity, and the tractability to be further optimized.
Organizational Affiliation:
Department of Chemical Biology and Therapeutics , St. Jude Children's Research Hospital , 262 Danny Thomas Place , Memphis , Tennessee 38105 , United States.